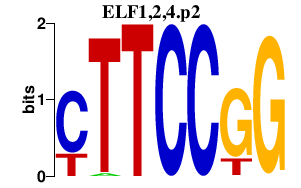
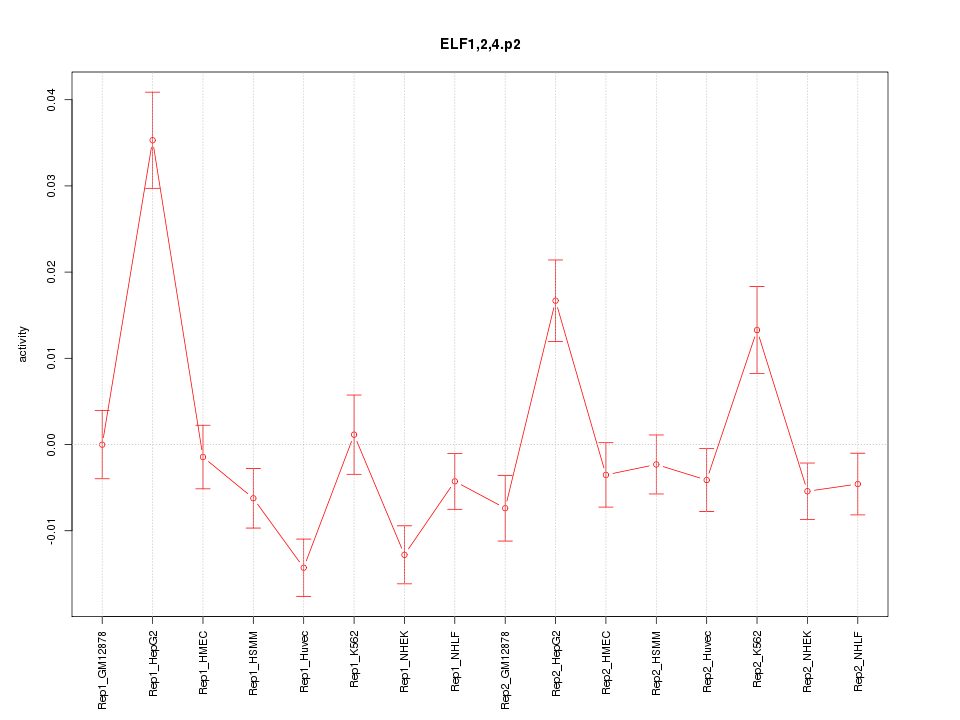
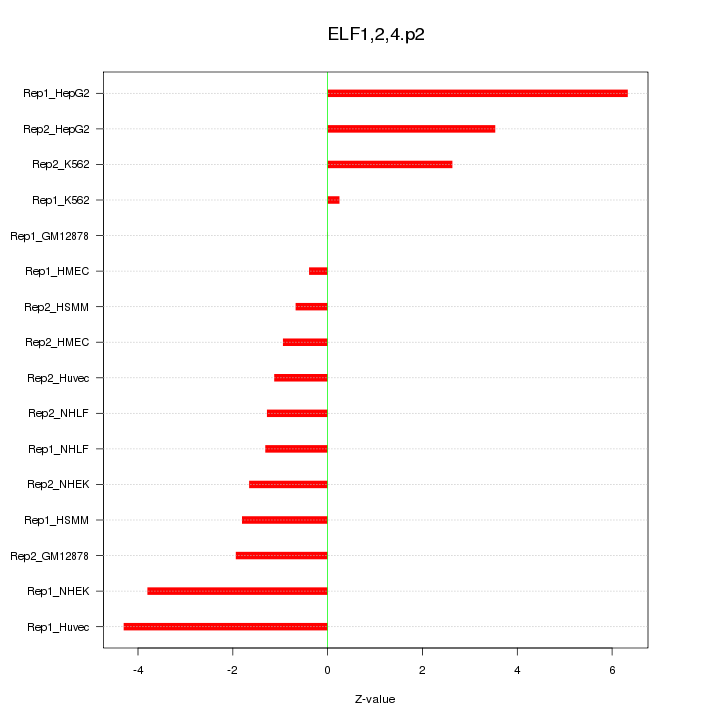
|
chr6_-_42527760
|
4.215
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr20_+_3138055
|
3.758
|
NM_033453
NM_181493
|
ITPA
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase)
|
|
chr2_-_73817935
|
3.477
|
NM_016058
|
TPRKB
|
TP53RK binding protein
|
|
chr14_+_22845851
|
3.384
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr2_+_198026470
|
3.262
|
NM_025147
|
COQ10B
|
coenzyme Q10 homolog B (S. cerevisiae)
|
|
chr2_-_45691887
|
3.139
|
NM_018079
|
SRBD1
|
S1 RNA binding domain 1
|
|
chr2_-_201461945
|
2.963
|
NM_032472
NM_130906
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3
|
|
chr2_-_207738858
|
2.841
|
NM_003709
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr20_+_33750645
|
2.701
|
NM_080748
|
ROMO1
|
reactive oxygen species modulator 1
|
|
chr20_+_60191455
|
2.691
|
NM_015666
|
GTPBP5
|
GTP binding protein 5 (putative)
|
|
chr10_-_51041156
|
2.669
|
NM_003631
|
AGAP8
LOC728407
PARG
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 8
poly(ADP-ribose) glycohydrolase pseudogene
poly (ADP-ribose) glycohydrolase
|
|
chr17_+_43328514
|
2.595
|
NM_003110
|
SP2
|
Sp2 transcription factor
|
|
chr2_-_54051290
|
2.595
|
NM_014614
|
PSME4
|
proteasome (prosome, macropain) activator subunit 4
|
|
chr20_+_18496068
|
2.574
|
|
LOC388789
|
hypothetical LOC388789
|
|
chr20_+_16658608
|
2.564
|
NM_003092
NM_198220
|
SNRPB2
|
small nuclear ribonucleoprotein polypeptide B
|
|
chr16_+_8799170
|
2.535
|
NM_000303
|
PMM2
|
phosphomannomutase 2
|
|
chr12_-_119116848
|
2.529
|
NM_006836
|
GCN1L1
|
GCN1 general control of amino-acid synthesis 1-like 1 (yeast)
|
|
chr2_-_27147995
|
2.494
|
NM_001134693
|
OST4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae)
|
|
chr6_-_119137923
|
2.465
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr16_+_2593351
|
2.430
|
|
LOC652276
|
hypothetical LOC652276
|
|
chr20_-_33750457
|
2.416
|
NM_001198989
NM_021100
|
NFS1
|
NFS1 nitrogen fixation 1 homolog (S. cerevisiae)
|
|
chr2_-_55349818
|
2.369
|
NM_001005369
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr16_-_8798973
|
2.346
|
NM_015421
|
TMEM186
|
transmembrane protein 186
|
|
chr20_-_49008445
|
2.338
|
NM_003859
|
DPM1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit
|
|
chr17_+_42355492
|
2.325
|
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr10_-_51293220
|
2.289
|
NM_006327
|
TIMM23
TIMM23B
|
translocase of inner mitochondrial membrane 23 homolog (yeast)
translocase of inner mitochondrial membrane 23 homolog B (yeast)
|
|
chr1_+_39229474
|
2.229
|
NM_001136275
NM_024595
|
AKIRIN1
|
akirin 1
|
|
chr16_-_11852862
|
2.205
|
NM_015659
|
RSL1D1
|
ribosomal L1 domain containing 1
|
|
chr17_-_39447817
|
2.203
|
NM_032376
|
TMEM101
|
transmembrane protein 101
|
|
chr1_-_28842103
|
2.167
|
NM_001135218
NM_005644
|
TAF12
|
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa
|
|
chr17_+_70520339
|
2.157
|
NM_001545
|
ICT1
|
immature colon carcinoma transcript 1
|
|
chr14_+_23982017
|
2.147
|
|
|
|
|
chr20_-_33198721
|
2.086
|
NM_001145025
NM_018217
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr16_-_19636918
|
2.083
|
NM_001012991
|
C16orf88
|
chromosome 16 open reading frame 88
|
|
chr14_+_34831294
|
2.081
|
NM_002791
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6
|
|
chr14_+_34831355
|
2.069
|
|
PSMA6
|
proteasome (prosome, macropain) subunit, alpha type, 6
|
|
chr2_+_201462294
|
2.035
|
NM_001136039
NM_001142355
NM_021824
|
NIF3L1
|
NIF3 NGG1 interacting factor 3-like 1 (S. pombe)
|
|
chr16_+_48865521
|
2.010
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr16_+_68938264
|
1.984
|
NM_018332
|
DDX19A
|
DEAD (Asp-Glu-Ala-As) box polypeptide 19A
|
|
chr14_-_21049204
|
1.974
|
NM_019852
|
METTL3
|
methyltransferase like 3
|
|
chr4_+_109761170
|
1.950
|
NM_000995
NM_033625
|
RPL34
|
ribosomal protein L34
|
|
chr2_+_113058470
|
1.934
|
NM_032309
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr2_-_37752731
|
1.908
|
NM_006449
|
CDC42EP3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr14_-_22634134
|
1.876
|
|
ACIN1
|
apoptotic chromatin condensation inducer 1
|
|
chr20_-_2592777
|
1.857
|
NM_006899
NM_174855
NM_174856
|
IDH3B
|
isocitrate dehydrogenase 3 (NAD+) beta
|
|
chr17_-_44377094
|
1.851
|
NM_007241
|
SNF8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae)
|
|
chr14_-_23981493
|
1.843
|
NM_020195
|
SDR39U1
|
short chain dehydrogenase/reductase family 39U, member 1
|
|
chr1_+_26358097
|
1.837
|
NM_024869
|
GRRP1
|
glycine/arginine rich protein 1
|
|
chr15_+_32304477
|
1.830
|
NM_016454
|
TMEM85
|
transmembrane protein 85
|
|
chr2_-_55349851
|
1.815
|
NM_002453
|
MTIF2
|
mitochondrial translational initiation factor 2
|
|
chr2_+_113058642
|
1.811
|
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5
|
|
chr17_+_64922420
|
1.810
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr12_+_12829807
|
1.805
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr20_+_49008754
|
1.796
|
NM_014484
|
MOCS3
|
molybdenum cofactor synthesis 3
|
|
chr6_-_167491305
|
1.788
|
NM_005299
|
GPR31
|
G protein-coupled receptor 31
|
|
chr20_-_33198555
|
1.782
|
|
EDEM2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr1_-_224993498
|
1.772
|
NM_002221
|
ITPKB
|
inositol 1,4,5-trisphosphate 3-kinase B
|
|
chr20_+_29598848
|
1.749
|
|
HM13
|
histocompatibility (minor) 13
|
|
chr20_+_58064360
|
1.747
|
NM_173644
|
C20orf197
|
chromosome 20 open reading frame 197
|
|
chr2_+_48521411
|
1.745
|
NM_001135629
NM_001193475
NM_152994
|
KLRAQ1
|
KLRAQ motif containing 1
|
|
chr14_+_23937766
|
1.727
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr20_-_5879036
|
1.721
|
NM_015939
|
TRMT6
|
tRNA methyltransferase 6 homolog (S. cerevisiae)
|
|
chr14_+_73423260
|
1.721
|
NM_021188
|
ZNF410
|
zinc finger protein 410
|
|
chr10_-_89567863
|
1.715
|
NM_032810
|
ATAD1
|
ATPase family, AAA domain containing 1
|
|
chr4_-_153820535
|
1.711
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr14_+_23771464
|
1.704
|
NM_001002000
NM_001002001
|
GMPR2
|
guanosine monophosphate reductase 2
|
|
chr6_-_169395972
|
1.702
|
NM_003247
|
THBS2
|
thrombospondin 2
|
|
chr6_-_169395847
|
1.699
|
|
THBS2
|
thrombospondin 2
|
|
chr6_+_26646548
|
1.691
|
NM_006353
|
HMGN4
|
high mobility group nucleosomal binding domain 4
|
|
chr2_+_183651531
|
1.663
|
NM_001142314
NM_080876
|
DUSP19
|
dual specificity phosphatase 19
|
|
chr2_+_220170804
|
1.662
|
NM_052902
|
STK11IP
|
serine/threonine kinase 11 interacting protein
|
|
chr14_-_29466562
|
1.651
|
NM_002742
|
PRKD1
|
protein kinase D1
|
|
chr3_-_172660545
|
1.650
|
NM_001161560
NM_001161561
NM_001161562
NM_001161563
NM_001161564
NM_001161565
NM_001161566
NM_015028
|
TNIK
|
TRAF2 and NCK interacting kinase
|
|
chr1_-_111308017
|
1.646
|
NM_001006945
NM_018372
|
C1orf103
|
chromosome 1 open reading frame 103
|
|
chr11_-_117628211
|
1.636
|
NM_198275
|
MPZL3
|
myelin protein zero-like 3
|
|
chr12_+_99118692
|
1.629
|
NM_022496
|
ACTR6
|
ARP6 actin-related protein 6 homolog (yeast)
|
|
chr2_-_74553285
|
1.624
|
NM_053050
|
MRPL53
|
mitochondrial ribosomal protein L53
|
|
chr21_+_42807052
|
1.624
|
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr1_+_42896634
|
1.611
|
NM_006347
|
PPIH
|
peptidylprolyl isomerase H (cyclophilin H)
|
|
chr7_-_44496703
|
1.610
|
NM_015332
|
NUDCD3
|
NudC domain containing 3
|
|
chr11_-_58102169
|
1.593
|
NM_001143995
|
LPXN
|
leupaxin
|
|
chr3_-_130600931
|
1.593
|
|
RPL32P3
|
ribosomal protein L32 pseudogene 3
|
|
chr6_-_30818198
|
1.592
|
NM_005803
|
FLOT1
|
flotillin 1
|
|
chr6_-_28327980
|
1.582
|
NM_019110
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4
|
|
chr2_-_1905508
|
1.582
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr16_-_15095590
|
1.566
|
NM_018427
|
RRN3
|
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae)
|
|
chr12_+_67366989
|
1.564
|
NM_020401
|
NUP107
|
nucleoporin 107kDa
|
|
chr1_-_23376860
|
1.562
|
|
LUZP1
|
leucine zipper protein 1
|
|
chr6_+_143813472
|
1.557
|
NM_003630
|
PEX3
|
peroxisomal biogenesis factor 3
|
|
chr14_+_74418345
|
1.551
|
NM_001933
|
DLST
|
dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex)
|
|
chr2_-_53867488
|
1.537
|
NM_016115
NM_145863
|
ASB3
|
ankyrin repeat and SOCS box containing 3
|
|
chr6_+_139136349
|
1.537
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr17_-_71174816
|
1.531
|
NM_004259
NM_001003715
NM_001003716
|
RECQL5
|
RecQ protein-like 5
|
|
chr20_+_43478120
|
1.523
|
NM_001184728
NM_001184729
NM_001184730
NM_015937
|
PIGT
|
phosphatidylinositol glycan anchor biosynthesis, class T
|
|
chr8_+_30133354
|
1.523
|
NM_006571
|
DCTN6
|
dynactin 6
|
|
chr1_+_22224571
|
1.522
|
|
HSPC157
|
hypothetical LOC29092
|
|
chr1_-_220829704
|
1.521
|
NM_005681
NM_139352
|
TAF1A
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa
|
|
chr14_+_23008742
|
1.521
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr12_+_70519753
|
1.509
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr10_-_18980512
|
1.506
|
NM_182543
|
NSUN6
|
NOP2/Sun domain family, member 6
|
|
chr12_+_118590039
|
1.504
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr2_+_176842368
|
1.495
|
NM_006554
|
MTX2
|
metaxin 2
|
|
chr19_-_51909416
|
1.494
|
NM_001079882
|
PRKD2
|
protein kinase D2
|
|
chr6_+_111409933
|
1.490
|
NM_032194
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae)
|
|
chr1_+_232575850
|
1.471
|
NM_001012985
|
C1orf31
|
chromosome 1 open reading frame 31
|
|
chr6_-_27548438
|
1.467
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chr3_+_30623376
|
1.460
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr6_+_30142910
|
1.459
|
NM_021959
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11
|
|
chr20_-_34005619
|
1.459
|
|
SCAND1
|
SCAN domain containing 1
|
|
chr2_+_32244406
|
1.444
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr8_+_59486376
|
1.441
|
NM_001077619
|
UBXN2B
|
UBX domain protein 2B
|
|
chr14_+_23008723
|
1.430
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr2_+_149119029
|
1.430
|
NM_015630
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila)
|
|
chr15_-_72952648
|
1.419
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr6_-_46997579
|
1.418
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr8_-_28403652
|
1.415
|
NM_172366
|
FBXO16
ZNF395
|
F-box protein 16
zinc finger protein 395
|
|
chr5_-_112658200
|
1.409
|
|
MCC
|
mutated in colorectal cancers
|
|
chr3_+_120495906
|
1.407
|
NM_020754
|
ARHGAP31
|
Rho GTPase activating protein 31
|
|
chr17_-_38229790
|
1.405
|
NM_003766
|
BECN1
|
beclin 1, autophagy related
|
|
chr3_+_3056307
|
1.404
|
NM_175612
|
CNTN4
|
contactin 4
|
|
chr6_+_144206165
|
1.404
|
NM_032860
|
LTV1
|
LTV1 homolog (S. cerevisiae)
|
|
chr2_+_65517316
|
1.402
|
|
FLJ16124
|
FLJ16124 protein
|
|
chr2_+_122124034
|
1.399
|
|
LOC254128
|
hypothetical LOC254128
|
|
chr1_+_144149794
|
1.397
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr20_-_44751208
|
1.395
|
|
TP53RK
|
TP53 regulating kinase
|
|
chr11_-_17055763
|
1.386
|
NM_001017
|
RPS13
|
ribosomal protein S13
|
|
chr6_-_34501769
|
1.385
|
NM_001014
|
RPS10
|
ribosomal protein S10
|
|
chr18_-_70415788
|
1.382
|
|
LOC400657
|
hypothetical LOC400657
|
|
chr2_-_20053256
|
1.380
|
NM_001006657
NM_020779
|
WDR35
|
WD repeat domain 35
|
|
chr17_-_53784494
|
1.367
|
NM_003168
|
SUPT4H1
|
suppressor of Ty 4 homolog 1 (S. cerevisiae)
|
|
chr6_+_144206202
|
1.358
|
|
LTV1
|
LTV1 homolog (S. cerevisiae)
|
|
chr2_+_69974581
|
1.356
|
NM_006857
|
SNRNP27
|
small nuclear ribonucleoprotein 27kDa (U4/U6.U5)
|
|
chr19_+_60838170
|
1.353
|
|
|
|
|
chr14_-_23781607
|
1.351
|
NM_001099274
NM_012461
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr2_+_96790298
|
1.346
|
NM_020184
|
CNNM4
|
cyclin M4
|
|
chr10_+_129735796
|
1.340
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr1_-_203867402
|
1.333
|
|
ELK4
|
ELK4, ETS-domain protein (SRF accessory protein 1)
|
|
chr19_-_425875
|
1.330
|
NM_182577
|
ODF3L2
|
outer dense fiber of sperm tails 3-like 2
|
|
chr17_-_44010543
|
1.330
|
NM_024015
|
HOXB4
|
homeobox B4
|
|
chr6_+_133177403
|
1.329
|
|
RPS12
|
ribosomal protein S12
|
|
chr7_+_102775309
|
1.328
|
NM_002803
|
PSMC2
|
proteasome (prosome, macropain) 26S subunit, ATPase, 2
|
|
chr14_-_23781328
|
1.328
|
|
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2
|
|
chr2_+_65308332
|
1.327
|
NM_001005386
NM_005722
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast)
|
|
chr17_+_42355482
|
1.321
|
NM_001012511
NM_004287
NM_054022
|
GOSR2
|
golgi SNAP receptor complex member 2
|
|
chr2_-_177836773
|
1.318
|
NM_001145412
NM_001145413
|
NFE2L2
|
nuclear factor (erythroid-derived 2)-like 2
|
|
chr20_+_21231921
|
1.312
|
NM_012255
|
XRN2
|
5'-3' exoribonuclease 2
|
|
chr2_-_152740387
|
1.312
|
NM_005843
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr7_+_141084591
|
1.310
|
NM_003143
|
SSBP1
|
single-stranded DNA binding protein 1
|
|
chr6_+_28301031
|
1.300
|
NM_006299
|
ZNF193
|
zinc finger protein 193
|
|
chr2_-_88707978
|
1.297
|
NM_004836
|
EIF2AK3
|
eukaryotic translation initiation factor 2-alpha kinase 3
|
|
chr20_-_13567529
|
1.292
|
NM_017714
|
TASP1
|
taspase, threonine aspartase, 1
|
|
chr16_-_3601580
|
1.292
|
NM_032444
|
SLX4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae)
|
|
chr2_+_53867571
|
1.288
|
NM_001127397
NM_001127398
NM_015701
|
ERLEC1
|
endoplasmic reticulum lectin 1
|
|
chr17_+_34162525
|
1.283
|
NM_002795
|
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3
|
|
chr8_-_57149588
|
1.274
|
NM_001023
NM_001146227
|
RPS20
|
ribosomal protein S20
|
|
chr20_-_10362755
|
1.265
|
NM_170784
|
MKKS
|
McKusick-Kaufman syndrome
|
|
chr4_-_39316856
|
1.252
|
NM_174921
|
C4orf34
|
chromosome 4 open reading frame 34
|
|
chr20_-_34005799
|
1.249
|
NM_016558
NM_033630
|
SCAND1
|
SCAN domain containing 1
|
|
chr17_-_26257341
|
1.248
|
NM_024683
|
C17orf42
|
chromosome 17 open reading frame 42
|
|
chr17_-_53950191
|
1.246
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr2_-_174538675
|
1.244
|
NM_001172712
NM_003111
|
SP3
|
Sp3 transcription factor
|
|
chr6_-_170735593
|
1.232
|
NM_002598
NM_144781
|
PDCD2
|
programmed cell death 2
|
|
chr17_+_35046843
|
1.227
|
NM_001165937
NM_001165938
NM_006804
|
STARD3
|
StAR-related lipid transfer (START) domain containing 3
|
|
chr2_+_230495439
|
1.224
|
NM_174899
|
FBXO36
|
F-box protein 36
|
|
chr20_-_472335
|
1.222
|
NM_001895
NM_177559
NM_177560
|
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide
|
|
chr3_+_144203061
|
1.217
|
NM_001080415
|
U2SURP
|
U2 snRNP-associated SURP domain containing
|
|
chr3_+_180548173
|
1.211
|
NM_033540
|
MFN1
|
mitofusin 1
|
|
chr16_+_24648446
|
1.210
|
NM_014494
|
TNRC6A
|
trinucleotide repeat containing 6A
|
|
chr2_+_239000395
|
1.210
|
|
ASB1
|
ankyrin repeat and SOCS box containing 1
|
|
chr18_+_31806585
|
1.207
|
NM_031446
|
C18orf21
|
chromosome 18 open reading frame 21
|
|
chr8_-_22582526
|
1.204
|
NM_018688
|
BIN3
|
bridging integrator 3
|
|
chr17_+_54642152
|
1.202
|
NM_018149
|
C17orf71
|
chromosome 17 open reading frame 71
|
|
chr2_+_27740254
|
1.200
|
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein
|
|
chr10_+_70609986
|
1.198
|
NM_003171
|
SUPV3L1
|
suppressor of var1, 3-like 1 (S. cerevisiae)
|
|
chr20_-_57040700
|
1.195
|
NM_006886
NM_001001977
|
ATP5E
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit
|
|
chr18_+_3252110
|
1.193
|
NM_033546
|
MYL12B
|
myosin, light chain 12B, regulatory
|
|
chr2_+_27739841
|
1.185
|
NM_018158
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein
|
|
chr2_-_230494862
|
1.185
|
NM_004238
|
TRIP12
|
thyroid hormone receptor interactor 12
|
|
chr5_-_159778621
|
1.184
|
NM_006425
|
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae)
|
|
chr17_+_38178999
|
1.180
|
|
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae)
|
|
chr6_+_149109841
|
1.174
|
NM_005715
|
UST
|
uronyl-2-sulfotransferase
|
|
chr7_-_27158726
|
1.172
|
|
|
|
|
chr20_+_30161173
|
1.169
|
|
TM9SF4
|
transmembrane 9 superfamily protein member 4
|
|
chr11_+_8660913
|
1.160
|
|
RPL27A
|
ribosomal protein L27a
|
|
chr11_-_77026406
|
1.153
|
NM_001293
|
CLNS1A
|
chloride channel, nucleotide-sensitive, 1A
|
|
chr20_+_47862782
|
1.152
|
|
SLC9A8
|
solute carrier family 9 (sodium/hydrogen exchanger), member 8
|
|
chr1_+_45926433
|
1.151
|
NM_016486
|
TMEM69
|
transmembrane protein 69
|
|
chr14_-_22127954
|
1.145
|
NM_001344
|
DAD1
|
defender against cell death 1
|
|
chr7_+_143683365
|
1.144
|
NM_005435
|
ARHGEF5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr1_+_45822246
|
1.142
|
NM_001195193
NM_002482
NM_152298
|
NASP
|
nuclear autoantigenic sperm protein (histone-binding)
|
|
chr20_+_5879297
|
1.141
|
NM_032485
NM_182802
|
MCM8
|
minichromosome maintenance complex component 8
|
|
chr4_+_152240182
|
1.139
|
NM_001006
|
RPS3A
|
ribosomal protein S3A
|
|
chr12_-_64810768
|
1.139
|
NM_032338
|
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia)
|
|
chr20_+_23279372
|
1.138
|
NM_013248
|
NXT1
|
NTF2-like export factor 1
|
|
chr12_-_48508411
|
1.133
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chr6_+_33530297
|
1.124
|
NM_152735
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
|
chr7_-_91713044
|
1.123
|
NM_004912
NM_001013406
NM_194454
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr20_+_18217120
|
1.120
|
NM_001083330
NM_003434
|
ZNF133
|
zinc finger protein 133
|
|
chr6_-_26767892
|
1.116
|
NM_024639
|
ZNF322A
|
zinc finger protein 322A
|
|
chr12_+_88268845
|
1.116
|
|
LOC100131490
|
hypothetical LOC100131490
|