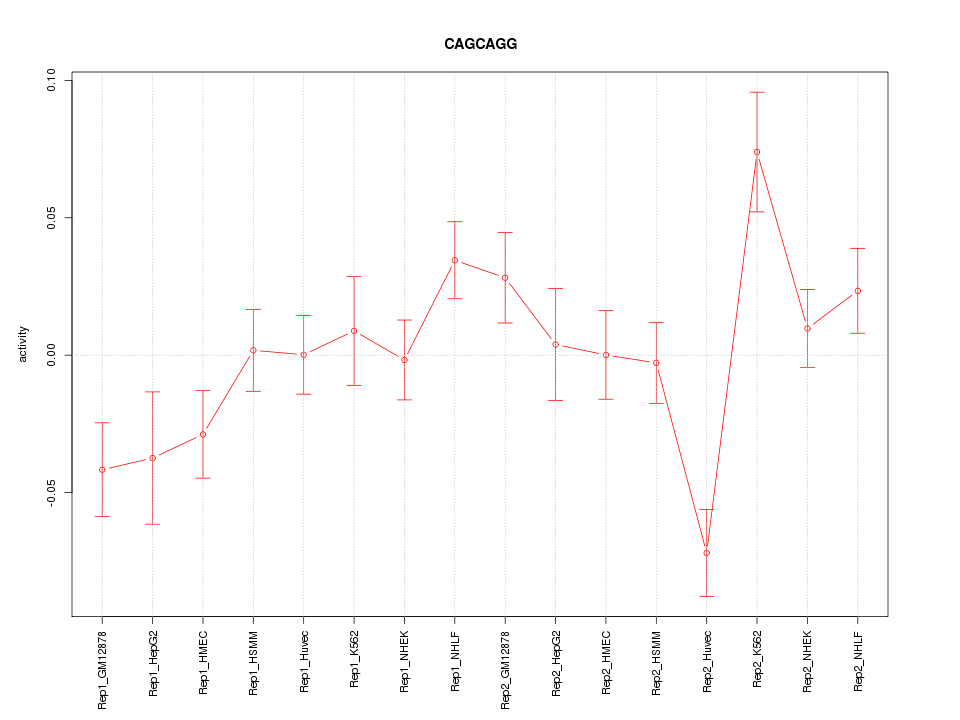
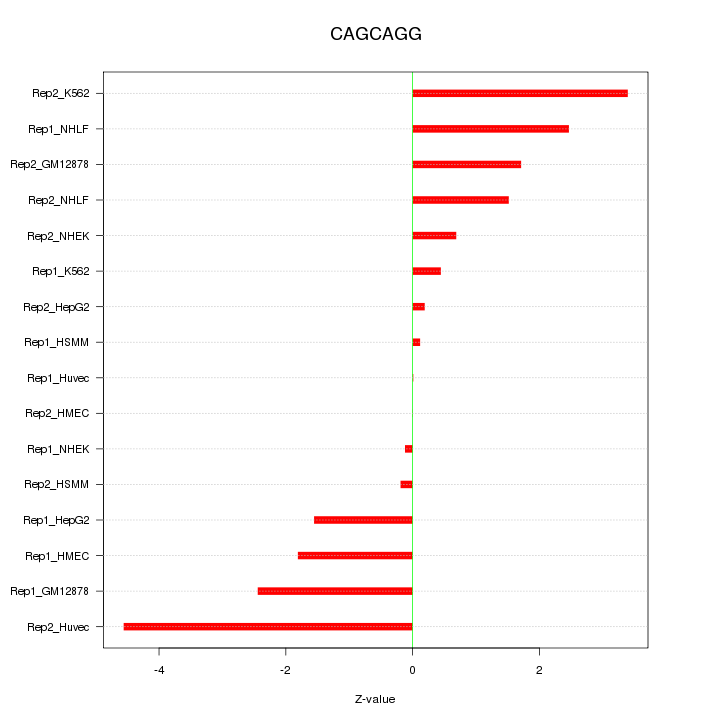
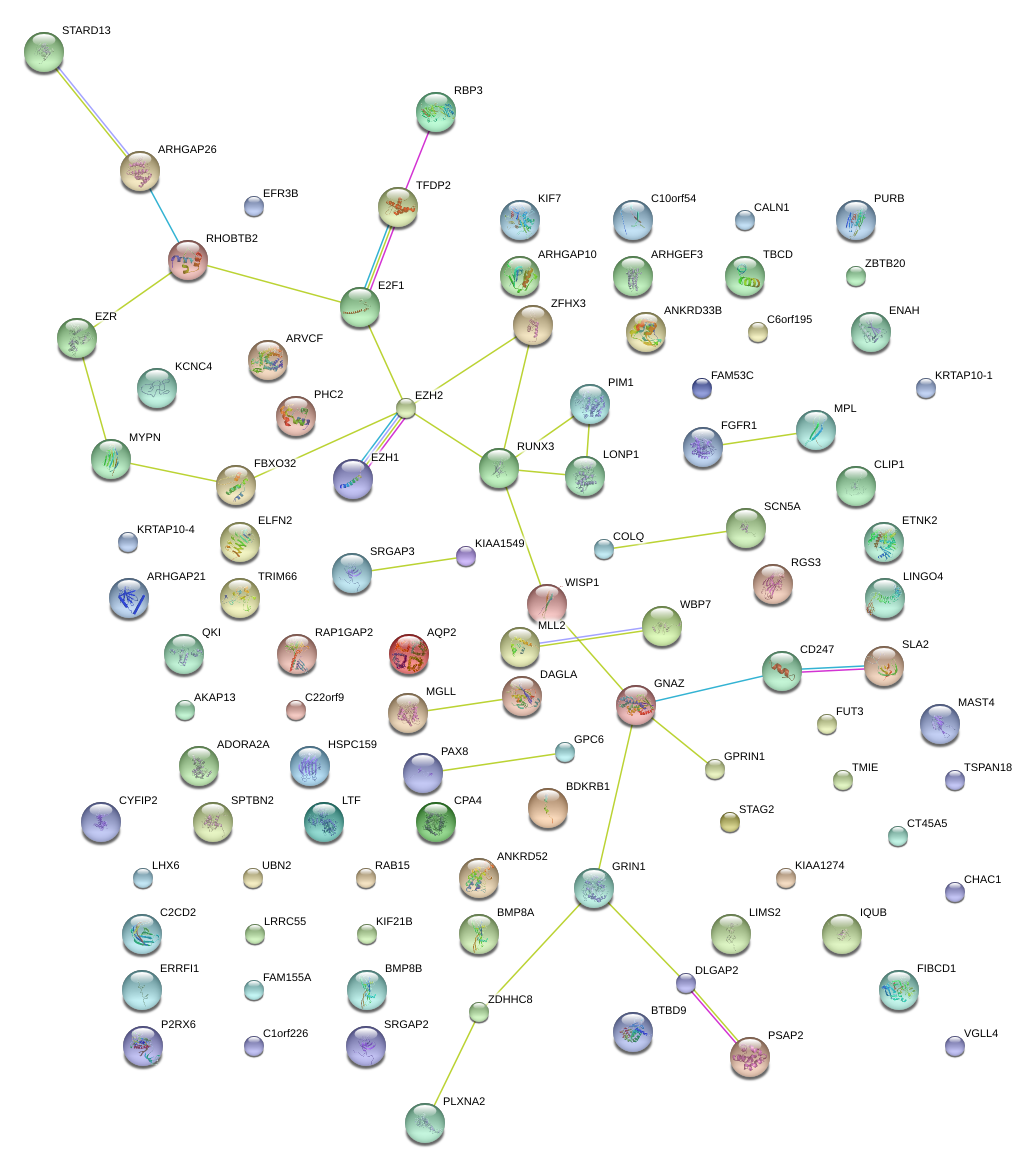
|
chr22_+_19699441
|
1.876
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr15_+_84021271
|
1.813
|
NM_144767
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr5_+_142130475
|
1.780
|
NM_001135608
NM_015071
|
ARHGAP26
|
Rho GTPase activating protein 26
|
|
chr11_+_61204392
|
1.689
|
NM_006133
|
DAGLA
|
diacylglycerol lipase, alpha
|
|
chr17_+_2646481
|
1.600
|
NM_001100398
NM_015085
|
RAP1GAP2
|
RAP1 GTPase activating protein 2
|
|
chr9_+_139153409
|
1.499
|
NM_000832
NM_001185090
NM_001185091
NM_007327
NM_021569
|
GRIN1
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1
|
|
chr22_+_21742364
|
1.437
|
NM_002073
|
GNAZ
|
guanine nucleotide binding protein (G protein), alpha z polypeptide
|
|
chr8_-_124613557
|
1.423
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr21_+_44818033
|
1.382
|
NM_198687
|
KRTAP10-4
|
keratin associated protein 10-4
|
|
chr1_-_206484258
|
1.359
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr3_-_56784634
|
1.358
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr1_+_110554836
|
1.340
|
NM_001039574
NM_004978
|
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4
|
|
chr22_-_18384251
|
1.298
|
NM_001670
|
ARVCF
|
armadillo repeat gene deleted in velocardiofacial syndrome
|
|
chr14_-_64508547
|
1.278
|
NM_198686
|
RAB15
|
RAB15, member RAS onocogene family
|
|
chr3_-_46481346
|
1.263
|
NM_002343
|
LTF
|
lactotransferrin
|
|
chr8_+_1436938
|
1.197
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr1_+_160618143
|
1.091
|
NM_001085375
|
C1orf226
|
chromosome 1 open reading frame 226
|
|
chr9_-_132804057
|
1.056
|
NM_032843
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr16_-_71650034
|
1.039
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr10_+_71908535
|
1.022
|
NM_014431
|
KIAA1274
|
KIAA1274
|
|
chrX_+_122922058
|
1.018
|
NM_001042749
|
STAG2
|
stromal antigen 2
|
|
chr1_-_202387767
|
1.011
|
NM_018208
|
ETNK2
|
ethanolamine kinase 2
|
|
chr22_+_23153529
|
1.009
|
NM_000675
|
ADORA2A
|
adenosine A2a receptor
|
|
chr21_-_42247007
|
1.000
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr8_+_22909305
|
0.998
|
NM_001160037
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr19_-_5802420
|
0.988
|
NM_000149
NM_001097639
NM_001097640
NM_001097641
|
FUT3
|
fucosyltransferase 3 (galactoside 3(4)-L-fucosyltransferase, Lewis blood group)
|
|
chr3_-_56810869
|
0.986
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr15_+_39032927
|
0.971
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr9_-_132804243
|
0.942
|
NM_001145106
|
FIBCD1
|
fibrinogen C domain containing 1
|
|
chr10_+_69539255
|
0.938
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr3_-_38666122
|
0.905
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr2_-_128138435
|
0.899
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr7_-_138316393
|
0.897
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr10_-_48010879
|
0.877
|
NM_002900
|
RBP3
|
retinol binding protein 3, interstitial
|
|
chr1_-_8008979
|
0.867
|
NM_018948
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr2_+_25118417
|
0.861
|
NM_014971
|
EFR3B
|
EFR3 homolog B (S. cerevisiae)
|
|
chr12_+_48630790
|
0.856
|
NM_000486
|
AQP2
|
aquaporin 2 (collecting duct)
|
|
chr6_+_163755658
|
0.849
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
quaking homolog, KH domain RNA binding (mouse)
|
|
chr1_-_199259447
|
0.841
|
NM_017596
|
KIF21B
|
kinesin family member 21B
|
|
chr13_-_107317460
|
0.833
|
NM_001080396
|
FAM155A
|
family with sequence similarity 155, member A
|
|
chr13_+_92677012
|
0.829
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr3_-_9266251
|
0.824
|
NM_001033117
NM_014850
|
SRGAP3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr1_-_223907283
|
0.820
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr9_-_124030770
|
0.806
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr5_+_156628929
|
0.799
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr7_-_44891471
|
0.785
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr2_-_113752957
|
0.783
|
NM_003466
NM_013952
NM_013953
NM_013992
|
PAX8
|
paired box 8
|
|
chr7_+_129720209
|
0.783
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr13_-_32658215
|
0.783
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr22_+_18499354
|
0.777
|
NM_001185024
NM_013373
|
ZDHHC8
|
zinc finger, DHHC-type containing 8
|
|
chr3_-_11736946
|
0.767
|
NM_014667
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr3_-_11598829
|
0.766
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr5_+_137701122
|
0.766
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chr15_-_87999528
|
0.759
|
NM_198525
|
KIF7
|
kinesin family member 7
|
|
chr3_-_15515382
|
0.753
|
NM_080538
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr11_+_56705796
|
0.750
|
NM_001005210
|
LRRC55
|
leucine rich repeat containing 55
|
|
chr17_-_38150551
|
0.748
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|
|
chr5_+_10617431
|
0.738
|
NM_001164440
|
ANKRD33B
|
ankyrin repeat domain 33B
|
|
chr8_-_38444398
|
0.734
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr12_-_54938385
|
0.726
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr8_-_38445508
|
0.721
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr8_+_134272463
|
0.719
|
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr3_-_115825742
|
0.709
|
NM_001164347
|
ZBTB20
|
zinc finger and BTB domain containing 20
|
|
chr22_-_36153449
|
0.702
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr21_-_44784499
|
0.701
|
NM_198691
|
KRTAP10-1
|
keratin associated protein 10-1
|
|
chr8_-_124622434
|
0.698
|
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr16_-_71639670
|
0.695
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr1_-_165754325
|
0.691
|
NM_000734
NM_198053
|
CD247
|
CD247 molecule
|
|
chr6_-_2580296
|
0.675
|
NM_152554
|
C6orf195
|
chromosome 6 open reading frame 195
|
|
chr10_-_73203202
|
0.674
|
NM_022153
|
C10orf54
|
chromosome 10 open reading frame 54
|
|
chr1_-_150044365
|
0.672
|
NM_001004432
|
LINGO4
|
leucine rich repeat and Ig domain containing 4
|
|
chr1_-_33613729
|
0.660
|
NM_198040
|
PHC2
|
polyhomeotic homolog 2 (Drosophila)
|
|
chr4_+_148872902
|
0.660
|
NM_024605
|
ARHGAP10
|
Rho GTPase activating protein 10
|
|
chr3_-_143230124
|
0.659
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr9_+_115367122
|
0.658
|
NM_134427
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_+_43576061
|
0.651
|
NM_005373
|
MPL
|
myeloproliferative leukemia virus oncogene
|
|
chr22_-_43987010
|
0.650
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr20_-_34707971
|
0.644
|
NM_032214
NM_175077
|
SLA2
|
Src-like-adaptor 2
|
|
chr5_+_66160359
|
0.643
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr6_-_38671741
|
0.641
|
NM_001172418
NM_152733
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr1_-_25164061
|
0.640
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr7_+_138566770
|
0.633
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr1_+_39729885
|
0.628
|
NM_181809
|
BMP8A
|
bone morphogenetic protein 8a
|
|
chr12_-_47735373
|
0.627
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr11_-_66245288
|
0.627
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr2_+_64534774
|
0.627
|
NM_014181
|
HSPC159
|
galectin-related protein
|
|
chr6_+_37245860
|
0.621
|
NM_002648
|
PIM1
|
pim-1 oncogene
|
|
chr11_+_44742551
|
0.619
|
NM_130783
|
TSPAN18
|
tetraspanin 18
|
|
chr8_+_22900863
|
0.619
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr11_-_8636958
|
0.617
|
NM_014818
|
TRIM66
|
tripartite motif containing 66
|
|
chr7_-_71515295
|
0.615
|
NM_031468
|
CALN1
|
calneuron 1
|
|
chr3_-_129024665
|
0.609
|
NM_001003794
|
MGLL
|
monoglyceride lipase
|
|
chr22_+_49459935
|
0.606
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr3_+_46717826
|
0.605
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr6_-_35804337
|
0.602
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr1_+_19842309
|
0.602
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr1_+_15123165
|
0.596
|
NM_001018000
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr2_-_128155829
|
0.594
|
NM_001161404
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr22_+_20316968
|
0.590
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chr6_-_170441493
|
0.589
|
NM_005618
|
DLL1
|
delta-like 1 (Drosophila)
|
|
chr2_+_176724358
|
0.588
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chrX_-_71268475
|
0.587
|
NM_001024455
|
RGAG4
|
retrotransposon gag domain containing 4
|
|
chr16_-_56077827
|
0.585
|
NM_018110
|
DOK4
|
docking protein 4
|
|
chr1_-_112083387
|
0.583
|
NM_019099
|
C1orf183
|
chromosome 1 open reading frame 183
|
|
chr6_+_90199521
|
0.581
|
NM_014942
|
ANKRD6
|
ankyrin repeat domain 6
|
|
chr2_+_128119908
|
0.581
|
NM_001161415
NM_001161416
NM_001161417
NM_005291
|
GPR17
|
G protein-coupled receptor 17
|
|
chr12_-_51200347
|
0.580
|
NM_000424
|
KRT5
|
keratin 5
|
|
chr1_+_165564904
|
0.578
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_36121382
|
0.577
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr3_-_170864111
|
0.567
|
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr1_-_172228673
|
0.565
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr6_-_38715901
|
0.565
|
NM_001099272
NM_052893
|
BTBD9
|
BTB (POZ) domain containing 9
|
|
chr9_+_129893948
|
0.563
|
NM_001006642
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25
|
|
chr17_-_6400470
|
0.559
|
NM_001165966
NM_031220
|
PITPNM3
|
PITPNM family member 3
|
|
chr5_+_125828685
|
0.556
|
NM_001146322
|
GRAMD3
|
GRAM domain containing 3
|
|
chr14_+_74968589
|
0.555
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr1_+_200698493
|
0.550
|
NM_032103
NM_032104
|
PPP1R12B
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr6_+_151688327
|
0.549
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr16_-_20271530
|
0.548
|
NM_001008389
NM_003361
|
UMOD
|
uromodulin
|
|
chr1_-_37752925
|
0.546
|
NM_022756
|
MEAF6
|
MYST/Esa1-associated factor 6
|
|
chr2_-_31659472
|
0.546
|
NM_000348
|
SRD5A2
|
steroid-5-alpha-reductase, alpha polypeptide 2 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 2)
|
|
chr1_-_200946038
|
0.544
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr1_+_152566899
|
0.539
|
NM_020452
|
ATP8B2
|
ATPase, class I, type 8B, member 2
|
|
chr22_-_19716837
|
0.538
|
NM_004173
|
SLC7A4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4
|
|
chr5_+_173247919
|
0.537
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr10_-_128199986
|
0.529
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr1_+_160733587
|
0.526
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr9_+_132874324
|
0.525
|
NM_006059
|
LAMC3
|
laminin, gamma 3
|
|
chr5_+_38439399
|
0.520
|
NM_182798
NM_182799
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr1_-_150065176
|
0.519
|
NM_001001523
|
RORC
|
RAR-related orphan receptor C
|
|
chr6_-_149847809
|
0.518
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr11_-_47502115
|
0.516
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr15_+_55455994
|
0.515
|
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr3_-_11585264
|
0.514
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr7_+_128293699
|
0.512
|
NM_001195150
|
LOC100130705
|
hypothetical protein LOC100130705
|
|
chr9_-_111230926
|
0.511
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr17_-_33753218
|
0.510
|
NM_001004334
|
GPR179
|
G protein-coupled receptor 179
|
|
chr18_+_53967544
|
0.508
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_+_135079461
|
0.508
|
NM_001167819
|
FHL1
|
four and a half LIM domains 1
|
|
chr17_+_4683280
|
0.502
|
NM_001024937
NM_015716
NM_153827
NM_170663
|
MINK1
|
misshapen-like kinase 1
|
|
chr19_+_19035792
|
0.499
|
NM_178526
|
SLC25A42
|
solute carrier family 25, member 42
|
|
chr18_+_45342398
|
0.498
|
NM_006033
|
LIPG
|
lipase, endothelial
|
|
chr3_+_14419087
|
0.498
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr1_+_200064904
|
0.498
|
NM_018085
|
IPO9
|
importin 9
|
|
chr15_-_50608296
|
0.497
|
NM_000259
NM_001142495
|
MYO5A
|
myosin VA (heavy chain 12, myoxin)
|
|
chr18_+_31488530
|
0.495
|
NM_020474
|
GALNT1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1)
|
|
chr1_+_27433593
|
0.494
|
NM_015023
|
WDTC1
|
WD and tetratricopeptide repeats 1
|
|
chr9_+_135314907
|
0.493
|
NM_001135775
NM_017586
|
C9orf7
|
chromosome 9 open reading frame 7
|
|
chr17_+_27838041
|
0.492
|
NM_003885
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr11_+_46339691
|
0.490
|
NM_001105540
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr13_+_95541093
|
0.488
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr5_+_176170041
|
0.488
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr5_-_88155360
|
0.486
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chrX_-_50573759
|
0.485
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr3_+_32123004
|
0.485
|
NM_015141
|
GPD1L
|
glycerol-3-phosphate dehydrogenase 1-like
|
|
chr5_-_159672060
|
0.483
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr14_+_54291255
|
0.483
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr22_+_18330049
|
0.483
|
NM_007310
|
COMT
|
catechol-O-methyltransferase
|
|
chr2_-_96145614
|
0.482
|
NM_000682
|
ADRA2B
|
adrenergic, alpha-2B-, receptor
|
|
chr13_+_112681626
|
0.480
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr16_-_70306198
|
0.479
|
NM_015020
|
PHLPP2
|
PH domain and leucine rich repeat protein phosphatase 2
|
|
chr12_-_52939636
|
0.479
|
NM_001127321
|
CBX5
|
chromobox homolog 5
|
|
chr9_-_135334003
|
0.475
|
NM_001145099
NM_017585
|
SLC2A6
|
solute carrier family 2 (facilitated glucose transporter), member 6
|
|
chr17_-_25587079
|
0.475
|
NM_001045
|
SLC6A4
|
solute carrier family 6 (neurotransmitter transporter, serotonin), member 4
|
|
chr1_+_19843261
|
0.471
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr1_-_52936611
|
0.470
|
NM_023077
|
C1orf163
|
chromosome 1 open reading frame 163
|
|
chr1_-_40903897
|
0.470
|
NM_014747
|
RIMS3
|
regulating synaptic membrane exocytosis 3
|
|
chr8_-_20156971
|
0.468
|
NM_021020
|
LZTS1
|
leucine zipper, putative tumor suppressor 1
|
|
chr11_-_112851051
|
0.466
|
NM_000795
NM_016574
|
DRD2
|
dopamine receptor D2
|
|
chr15_-_63457378
|
0.465
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chrX_+_49013243
|
0.465
|
NM_001184745
NM_033215
|
PPP1R3F
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr21_-_32879617
|
0.464
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr19_+_10402334
|
0.464
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr11_+_8016755
|
0.464
|
NM_003320
|
TUB
|
tubby homolog (mouse)
|
|
chr20_-_29921461
|
0.461
|
NM_001012644
|
DUSP15
|
dual specificity phosphatase 15
|
|
chr3_+_52788976
|
0.460
|
NM_001166435
NM_001166436
|
ITIH1
|
inter-alpha (globulin) inhibitor H1
|
|
chr20_+_34635362
|
0.457
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr2_+_26768894
|
0.452
|
NM_002246
|
KCNK3
|
potassium channel, subfamily K, member 3
|
|
chr3_-_57088332
|
0.451
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr12_-_48523123
|
0.448
|
NM_181708
|
BCDIN3D
|
BCDIN3 domain containing
|
|
chr11_-_76058542
|
0.448
|
NM_005512
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr5_-_141238127
|
0.448
|
NM_002587
NM_032420
|
PCDH1
|
protocadherin 1
|
|
chr18_+_13610866
|
0.447
|
NM_004338
NM_181483
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr1_-_200879203
|
0.445
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr11_+_46325457
|
0.443
|
NM_003646
|
DGKZ
|
diacylglycerol kinase, zeta
|
|
chr20_-_43163021
|
0.443
|
NM_002251
|
KCNS1
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1
|
|
chr15_+_65217412
|
0.443
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr5_-_56283672
|
0.441
|
NM_152622
|
MIER3
|
mesoderm induction early response 1, family member 3
|
|
chr12_+_1799693
|
0.439
|
NM_001039029
NM_001163925
NM_001163926
|
LRTM2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr9_+_130883199
|
0.439
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr1_-_45444812
|
0.437
|
NM_020883
|
ZSWIM5
|
zinc finger, SWIM-type containing 5
|
|
chr21_-_43671355
|
0.436
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr12_-_78831821
|
0.436
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory (inhibitor) subunit 12A
|
|
chr22_+_19601619
|
0.436
|
NM_005207
|
CRKL
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr3_+_39484073
|
0.435
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr1_-_32940943
|
0.433
|
NM_001161708
NM_030786
|
SYNC
|
syncoilin, intermediate filament protein
|
|
chr1_+_6407403
|
0.430
|
NM_031475
|
ESPN
|
espin
|
|
chr5_+_38481392
|
0.430
|
NM_182801
|
EGFLAM
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr12_-_51114372
|
0.426
|
NM_004693
|
KRT75
|
keratin 75
|
|
chr3_+_42519098
|
0.425
|
NM_004624
|
VIPR1
|
vasoactive intestinal peptide receptor 1
|