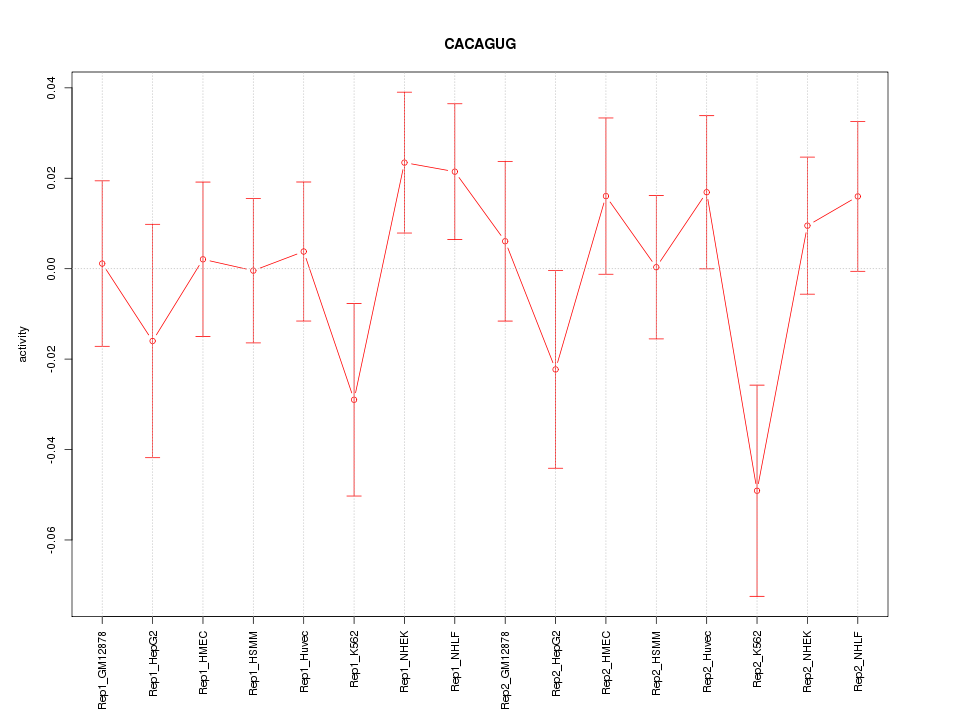
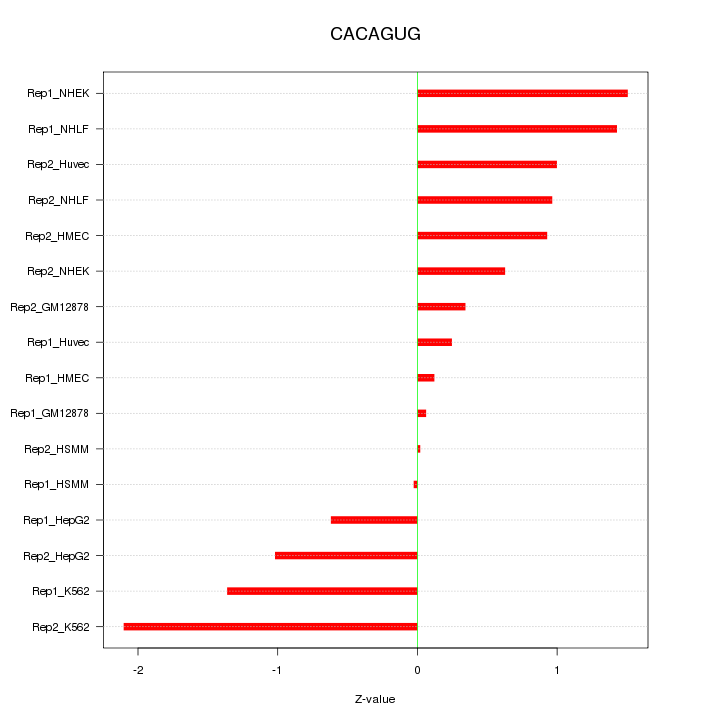
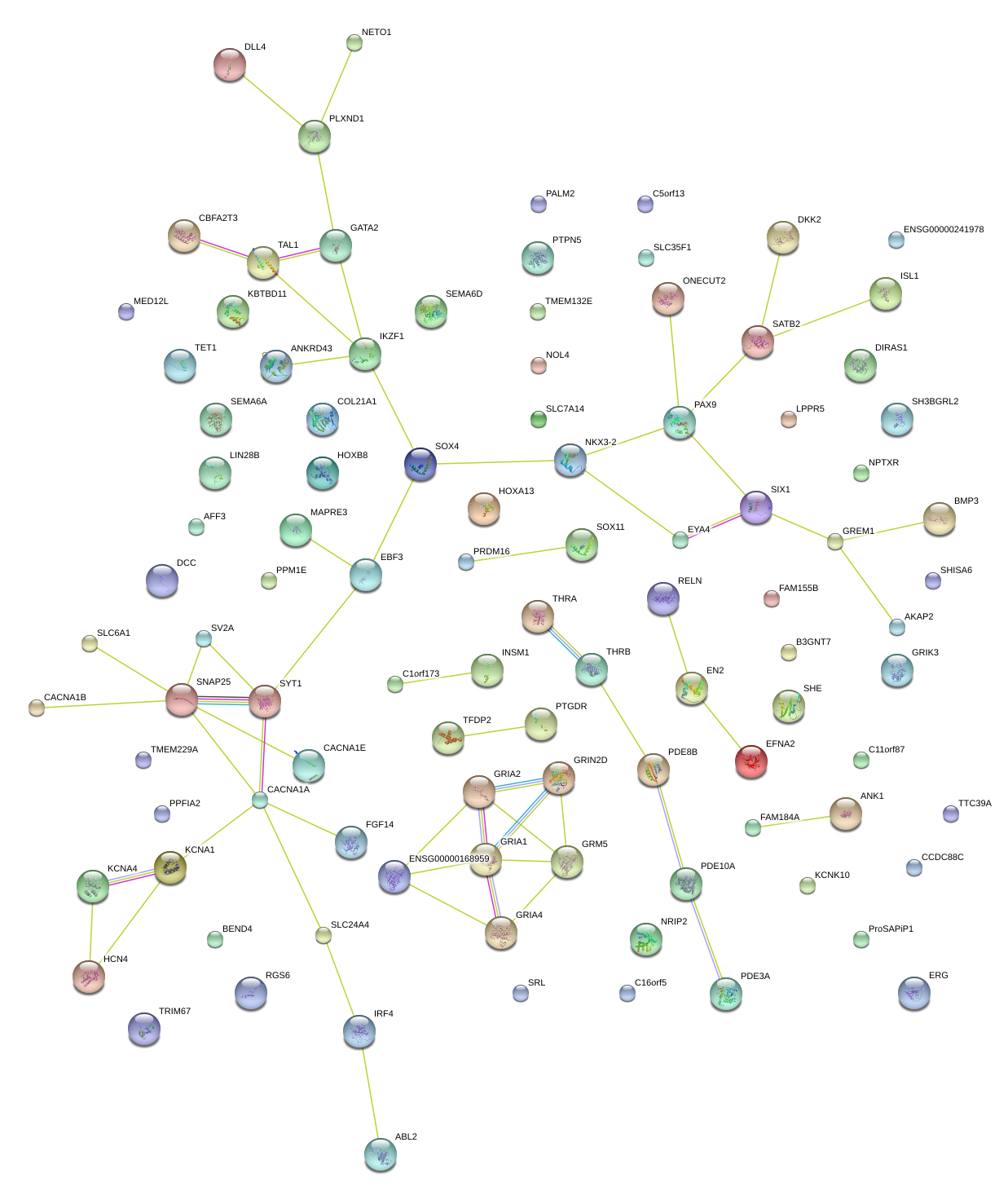
|
chr4_-_108176901
|
2.691
|
NM_014421
|
DKK2
|
dickkopf homolog 2 (Xenopus laevis)
|
|
chr4_-_41849651
|
2.453
|
NM_001159547
NM_207406
|
BEND4
|
BEN domain containing 4
|
|
chr18_+_53253775
|
2.407
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr3_-_129694717
|
2.293
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr5_+_132176921
|
2.262
|
NM_175873
|
ANKRD43
|
ankyrin repeat domain 43
|
|
chr7_-_28964553
|
2.210
|
NM_014817
|
TRIL
|
TLR4 interactor with leucine-rich repeats
|
|
chr3_-_171786468
|
2.190
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr14_+_91859904
|
2.004
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr17_-_44047299
|
1.899
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr15_+_30797466
|
1.851
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr7_+_154943466
|
1.769
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr18_-_68685789
|
1.755
|
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr22_-_37569962
|
1.712
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr12_-_80677223
|
1.672
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr21_-_38955487
|
1.648
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr4_-_13155135
|
1.640
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr16_-_87535106
|
1.551
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr1_-_148155986
|
1.525
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr2_+_5750229
|
1.506
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr7_-_103417198
|
1.496
|
NM_005045
NM_173054
|
RELN
|
reelin
|
|
chr18_-_30057512
|
1.488
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr14_+_91859289
|
1.474
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr5_-_115938305
|
1.440
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr19_+_53589943
|
1.438
|
NM_000836
|
GRIN2D
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D
|
|
chr3_-_129689394
|
1.432
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr5_-_111340421
|
1.408
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_50314801
|
1.404
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr14_+_51804180
|
1.386
|
NM_000953
|
PTGDR
|
prostaglandin D2 receptor (DP)
|
|
chr2_-_200044233
|
1.362
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr18_+_48120539
|
1.359
|
NM_005215
|
DCC
|
deleted in colorectal carcinoma
|
|
chr14_+_36196523
|
1.349
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr10_+_69990050
|
1.348
|
NM_030625
|
TET1
|
tet oncogene 1
|
|
chr2_-_200038055
|
1.343
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr11_-_88436463
|
1.338
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr19_-_13478037
|
1.328
|
NM_000068
NM_001127221
NM_001127222
NM_001174080
NM_023035
|
CACNA1A
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr13_-_101366995
|
1.312
|
NM_004115
|
FGF14
|
fibroblast growth factor 14
|
|
chr17_+_54188230
|
1.312
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr3_-_130807865
|
1.311
|
NM_015103
|
PLXND1
|
plexin D1
|
|
chr1_-_37272295
|
1.308
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr14_-_87863003
|
1.296
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr17_+_11085464
|
1.293
|
NM_001173461
NM_001173462
NM_207386
|
SHISA6
|
shisa homolog 6 (Xenopus laevis)
|
|
chr18_-_30056431
|
1.283
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr1_-_152741148
|
1.269
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr6_-_165995510
|
1.260
|
NM_001130690
NM_006661
|
PDE10A
|
phosphodiesterase 10A
|
|
chr19_-_2672337
|
1.249
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr1_+_2975590
|
1.248
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr6_+_80397690
|
1.241
|
NM_031469
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2
|
|
chrX_+_68641802
|
1.223
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr1_-_99242765
|
1.221
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr14_+_91858677
|
1.209
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr15_+_45263517
|
1.201
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr20_-_3097070
|
1.174
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr11_+_104986009
|
1.167
|
NM_000829
NM_001077243
NM_001077244
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr11_-_18769703
|
1.164
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr6_-_119441458
|
1.159
|
NM_024581
|
FAM184A
|
family with sequence similarity 184, member A
|
|
chr20_+_10147455
|
1.147
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr9_+_111442888
|
1.141
|
NM_001037293
|
PALM2
|
paralemmin 2
|
|
chr12_+_77781903
|
1.097
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr11_-_29995063
|
1.081
|
NM_002233
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4
|
|
chr2_+_231968578
|
1.078
|
NM_145236
|
B3GNT7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7
|
|
chr6_+_133604181
|
1.039
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr3_-_129690056
|
1.033
|
NM_001145661
|
GATA2
|
GATA binding protein 2
|
|
chr6_+_336738
|
1.014
|
NM_001195286
NM_002460
|
IRF4
|
interferon regulatory factor 4
|
|
chr6_-_56220274
|
1.003
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr1_-_51560469
|
0.995
|
NM_001080494
|
TTC39A
|
tetratricopeptide repeat domain 39A
|
|
chr3_+_11009419
|
0.992
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr12_+_20413445
|
0.990
|
NM_000921
|
PDE3A
|
phosphodiesterase 3A, cGMP-inhibited
|
|
chr15_+_39008822
|
0.980
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr11_+_108798047
|
0.963
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr14_+_71469538
|
0.955
|
NM_004296
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr14_-_90953883
|
0.954
|
NM_001080414
|
CCDC88C
|
coiled-coil domain containing 88C
|
|
chr1_-_47468029
|
0.953
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr8_+_1909437
|
0.949
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr3_-_24511180
|
0.943
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr17_+_29931880
|
0.939
|
NM_207313
|
TMEM132E
|
transmembrane protein 132E
|
|
chr12_-_2814410
|
0.936
|
NM_031474
|
NRIP2
|
nuclear receptor interacting protein 2
|
|
chr7_-_123460722
|
0.923
|
NM_001136002
|
TMEM229A
|
transmembrane protein 229A
|
|
chr20_+_20296744
|
0.918
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr19_+_1236766
|
0.910
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr2_-_100088224
|
0.909
|
NM_001025108
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr14_-_87859284
|
0.907
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr1_+_229365296
|
0.904
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr15_-_71448657
|
0.904
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr1_-_74912009
|
0.890
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr14_-_60185659
|
0.890
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr6_+_118335381
|
0.882
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr12_+_4889309
|
0.877
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr5_+_50714714
|
0.874
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr6_+_105511615
|
0.873
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr3_+_152287365
|
0.848
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr1_-_177378801
|
0.844
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr5_+_76542461
|
0.832
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr8_-_41873401
|
0.826
|
NM_001142446
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr10_-_131652007
|
0.819
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr16_-_4232053
|
0.811
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr7_-_27206249
|
0.803
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr16_-_4528739
|
0.803
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr4_+_82171142
|
0.801
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr3_-_143201878
|
0.797
|
NM_001178140
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr10_-_7493418
|
0.797
|
NM_001029880
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr13_+_112670757
|
0.797
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_-_36153449
|
0.785
|
NM_052906
|
ELFN2
|
extracellular leucine-rich repeat and fibronectin type III domain containing 2
|
|
chr9_+_102830851
|
0.775
|
NM_207299
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr7_-_27180398
|
0.764
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr1_+_38033666
|
0.748
|
NM_152496
|
MANEAL
|
mannosidase, endo-alpha-like
|
|
chr3_+_54131731
|
0.746
|
NM_018398
|
CACNA2D3
|
calcium channel, voltage-dependent, alpha 2/delta subunit 3
|
|
chr14_-_56346849
|
0.731
|
NM_021728
|
OTX2
|
orthodenticle homeobox 2
|
|
chr8_-_10625352
|
0.729
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr1_+_226261374
|
0.728
|
NM_033131
|
WNT3A
|
wingless-type MMTV integration site family, member 3A
|
|
chr3_+_23961739
|
0.726
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr21_+_32167175
|
0.717
|
NM_014586
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr2_-_119322228
|
0.714
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr12_-_122160927
|
0.709
|
NM_020845
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2
|
|
chr12_+_108636569
|
0.706
|
NM_032829
|
C12orf34
|
chromosome 12 open reading frame 34
|
|
chr20_-_41251878
|
0.706
|
NM_007050
NM_133170
|
PTPRT
|
protein tyrosine phosphatase, receptor type, T
|
|
chr1_-_182273451
|
0.703
|
NM_015101
|
GLT25D2
|
glycosyltransferase 25 domain containing 2
|
|
chr6_-_3390197
|
0.702
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr3_-_13089547
|
0.697
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr16_+_65436341
|
0.681
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chr1_+_158636987
|
0.675
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr13_-_24643778
|
0.670
|
NM_152704
NM_199138
|
FAM123A
|
family with sequence similarity 123A
|
|
chr17_+_12509931
|
0.669
|
NM_001146312
NM_153604
|
MYOCD
|
myocardin
|
|
chr14_-_56342097
|
0.651
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chr1_+_26609819
|
0.634
|
NM_024674
|
LIN28A
|
lin-28 homolog A (C. elegans)
|
|
chr10_-_91395194
|
0.634
|
NM_148977
|
PANK1
|
pantothenate kinase 1
|
|
chr9_+_34948171
|
0.634
|
NM_015297
|
KIAA1045
|
KIAA1045
|
|
chrX_-_79951838
|
0.629
|
NM_153252
|
BRWD3
|
bromodomain and WD repeat domain containing 3
|
|
chr6_-_33268177
|
0.624
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr19_+_19183763
|
0.623
|
NM_004386
|
NCAN
|
neurocan
|
|
chr12_+_53241365
|
0.618
|
NM_001165975
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr6_+_14033402
|
0.615
|
NM_001165033
|
RNF182
|
ring finger protein 182
|
|
chr22_-_27405708
|
0.613
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chrX_+_104953187
|
0.605
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr5_+_7449030
|
0.605
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr11_+_14621844
|
0.595
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr13_-_47899043
|
0.592
|
NM_001162497
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr5_-_179712920
|
0.588
|
NM_005110
|
GFPT2
|
glutamine-fructose-6-phosphate transaminase 2
|
|
chr2_-_158162271
|
0.587
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr7_+_94377145
|
0.576
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr12_+_77782579
|
0.576
|
NM_005639
|
SYT1
|
synaptotagmin I
|
|
chr9_-_23816062
|
0.576
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr3_+_36396948
|
0.573
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr9_-_23811842
|
0.569
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr8_-_41641881
|
0.559
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr11_-_88420687
|
0.558
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr10_-_7491204
|
0.554
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr2_+_210344945
|
0.541
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chr9_-_23811477
|
0.532
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chrX_-_135691168
|
0.530
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chrX_-_54401161
|
0.528
|
NM_001002838
NM_020922
|
WNK3
|
WNK lysine deficient protein kinase 3
|
|
chrX_-_40479718
|
0.521
|
NM_004229
|
MED14
|
mediator complex subunit 14
|
|
chr11_-_35397675
|
0.511
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr16_-_4528380
|
0.511
|
NM_001199054
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chr8_+_16929116
|
0.507
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr3_-_131081910
|
0.503
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr12_-_61614930
|
0.498
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr4_+_114190233
|
0.498
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr1_+_219119358
|
0.496
|
NM_021958
|
HLX
|
H2.0-like homeobox
|
|
chr21_-_27261276
|
0.495
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr3_-_144164867
|
0.483
|
NM_198504
|
PAQR9
|
progestin and adipoQ receptor family member IX
|
|
chr7_+_98084531
|
0.483
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr19_-_7244876
|
0.476
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr2_-_50428395
|
0.475
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr19_+_61846338
|
0.473
|
NM_001193628
|
LOC147670
|
hypothetical protein LOC147670
|
|
chr2_+_228045121
|
0.469
|
NM_001135187
NM_001135188
NM_001135189
NM_004504
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr18_-_29882555
|
0.466
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr22_+_40286957
|
0.461
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chrX_-_110542040
|
0.458
|
NM_178151
NM_001195553
NM_178152
NM_178153
|
DCX
|
doublecortin
|
|
chr10_+_102881050
|
0.456
|
NM_001195517
NM_005521
|
TLX1
|
T-cell leukemia homeobox 1
|
|
chr12_+_111980044
|
0.454
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr20_+_48032918
|
0.452
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr1_-_153098811
|
0.452
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr14_+_104337923
|
0.451
|
NM_001137601
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr17_-_29930431
|
0.441
|
NM_207454
|
C17orf102
|
chromosome 17 open reading frame 102
|
|
chr13_-_101852028
|
0.437
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr22_+_44925162
|
0.433
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr9_+_128416542
|
0.428
|
NM_001174146
NM_001174147
NM_002316
|
LMX1B
|
LIM homeobox transcription factor 1, beta
|
|
chr1_+_213323182
|
0.428
|
NM_001017425
NM_014217
|
KCNK2
|
potassium channel, subfamily K, member 2
|
|
chrX_+_16714452
|
0.427
|
NM_001168683
NM_018360
|
TXLNG
|
taxilin gamma
|
|
chr8_+_77756061
|
0.423
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr1_-_200946038
|
0.421
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr8_-_22606653
|
0.420
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr11_+_119616112
|
0.418
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chr17_+_1906262
|
0.412
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr3_-_13436802
|
0.409
|
NM_024923
|
NUP210
|
nucleoporin 210kDa
|
|
chr1_-_153109349
|
0.405
|
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr11_-_84312085
|
0.397
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr12_+_53699995
|
0.397
|
NM_021191
|
NEUROD4
|
neurogenic differentiation 4
|
|
chr4_-_174687931
|
0.394
|
NM_021973
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr3_+_141136691
|
0.391
|
NM_022131
|
CLSTN2
|
calsyntenin 2
|
|
chr6_-_88932485
|
0.389
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr4_-_53217513
|
0.387
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chrX_-_24943985
|
0.387
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chrX_+_70232362
|
0.386
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr16_-_67039874
|
0.383
|
NM_018667
|
SMPD3
|
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II)
|
|
chr20_+_24397834
|
0.379
|
NM_024893
|
TMEM90B
|
transmembrane protein 90B
|
|
chr14_+_76634323
|
0.377
|
NM_033426
|
KIAA1737
|
KIAA1737
|
|
chrX_-_129230434
|
0.377
|
NM_017666
|
ZNF280C
|
zinc finger protein 280C
|
|
chr12_+_76749199
|
0.376
|
NM_014903
|
NAV3
|
neuron navigator 3
|
|
chr9_-_92444886
|
0.374
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|