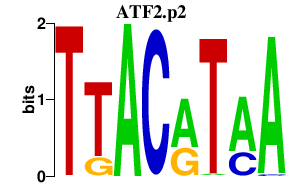
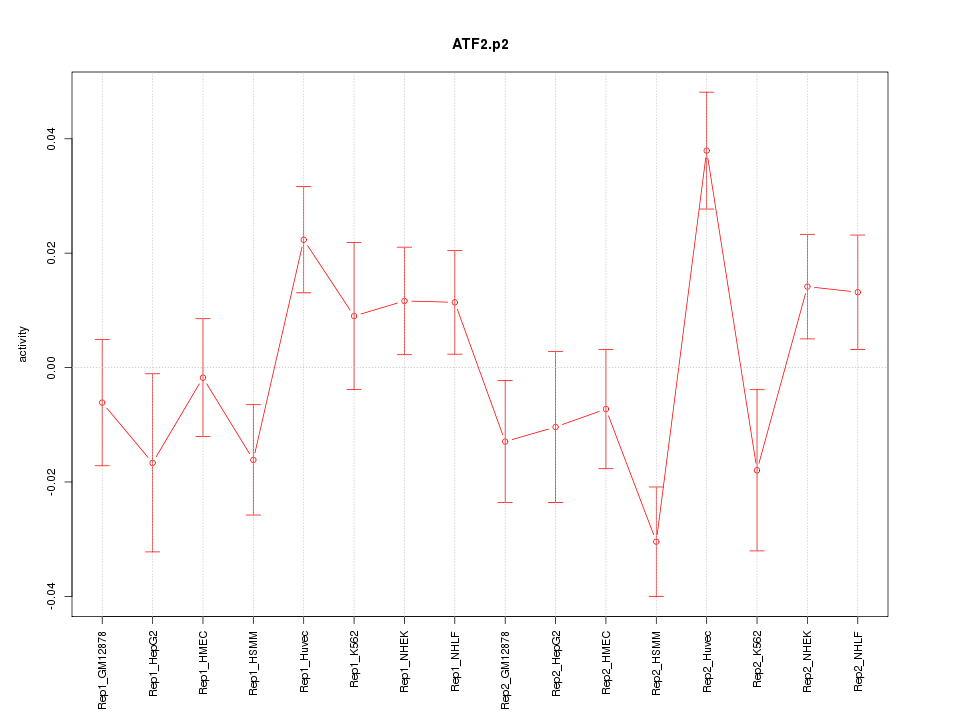
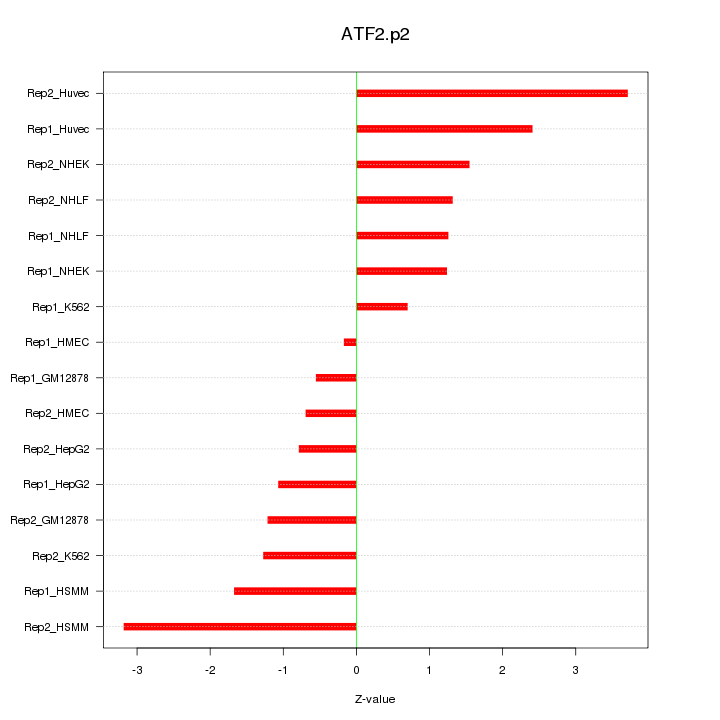
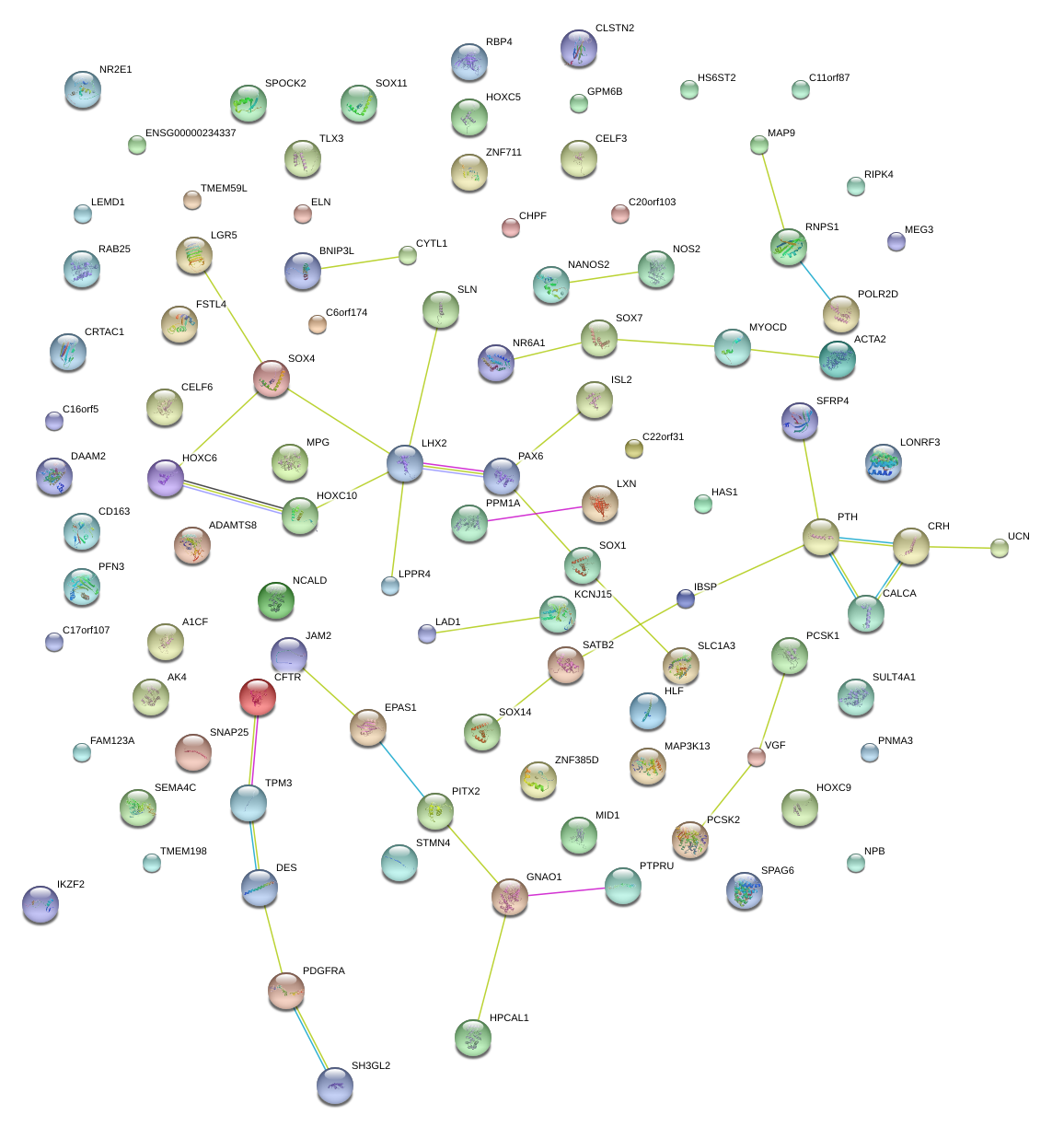
|
chr4_-_111777651
|
2.919
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr2_-_27384617
|
2.815
|
NM_003353
|
UCN
|
urocortin
|
|
chr4_-_111777584
|
2.723
|
|
PITX2
|
paired-like homeodomain 2
|
|
chr2_+_220116986
|
2.136
|
NM_001005209
|
TMEM198
|
transmembrane protein 198
|
|
chr15_+_74416156
|
2.006
|
NM_145805
|
ISL2
|
ISL LIM homeobox 2
|
|
chr5_-_176760242
|
1.974
|
NM_001029886
|
PFN3
|
profilin 3
|
|
chr2_+_220117009
|
1.949
|
|
TMEM198
|
transmembrane protein 198
|
|
chr15_+_74416129
|
1.916
|
|
ISL2
|
ISL LIM homeobox 2
|
|
chr5_+_140747586
|
1.898
|
NM_003736
NM_032098
|
PCDHGB4
|
protocadherin gamma subfamily B, 4
|
|
chr11_-_31796061
|
1.877
|
NM_001127612
|
PAX6
|
paired box 6
|
|
chr22_-_27787793
|
1.863
|
NM_015370
|
C22orf31
|
chromosome 22 open reading frame 31
|
|
chr19_+_18584666
|
1.755
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr12_+_52665196
|
1.731
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr2_-_96899396
|
1.638
|
NM_017789
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr12_+_70119785
|
1.629
|
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr10_+_22674379
|
1.603
|
NM_012443
NM_172242
|
SPAG6
|
sperm associated antigen 6
|
|
chr1_+_29435534
|
1.592
|
NM_001195001
NM_005704
NM_133177
NM_133178
|
PTPRU
|
protein tyrosine phosphatase, receptor type, U
|
|
chr9_+_125813672
|
1.443
|
NM_004789
|
LHX2
|
LIM homeobox 2
|
|
chrX_+_117992603
|
1.423
|
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr6_-_127822227
|
1.401
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chrX_-_13866565
|
1.399
|
NM_001001994
|
GPM6B
|
glycoprotein M6B
|
|
chr6_+_108593907
|
1.376
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr5_-_95794416
|
1.366
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr10_-_95350949
|
1.302
|
NM_006744
|
RBP4
|
retinol binding protein 4, plasma
|
|
chr21_-_42060266
|
1.275
|
NM_020639
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr11_-_107087943
|
1.251
|
NM_003063
|
SLN
|
sarcolipin
|
|
chr7_-_37923040
|
1.220
|
NM_003014
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr3_-_21767819
|
1.218
|
NM_024697
|
ZNF385D
|
zinc finger protein 385D
|
|
chr21_-_42060334
|
1.213
|
|
RIPK4
|
receptor-interacting serine-threonine kinase 4
|
|
chr12_+_52680143
|
1.189
|
NM_006897
|
HOXC9
|
homeobox C9
|
|
chr17_+_50697319
|
1.184
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr1_+_65385799
|
1.173
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr4_-_187884721
|
1.171
|
|
|
|
|
chr11_+_108798047
|
1.164
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chrX_+_84385649
|
1.152
|
NM_021998
|
ZNF711
|
zinc finger protein 711
|
|
chr2_+_5750229
|
1.152
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr6_+_39868116
|
1.139
|
|
|
|
|
chr22_-_42589543
|
1.129
|
NM_014351
|
SULT4A1
|
sulfotransferase family 4A, member 1
|
|
chr11_-_13474101
|
1.098
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr16_-_4528739
|
1.076
|
NM_001199055
NM_001199056
NM_013399
|
C16orf5
|
chromosome 16 open reading frame 5
|
|
chrX_-_10605726
|
1.069
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr14_+_59782222
|
1.062
|
NM_177952
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A
|
|
chr5_-_132976094
|
1.056
|
|
FSTL4
|
follistatin-like 4
|
|
chr17_+_50697355
|
1.048
|
|
HLF
|
hepatic leukemia factor
|
|
chr20_+_10147455
|
1.024
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr16_-_53520208
|
1.017
|
|
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding)
|
|
chr5_-_132976121
|
1.016
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr9_+_125813849
|
1.009
|
|
LHX2
|
LIM homeobox 2
|
|
chr6_+_39868126
|
0.994
|
|
DAAM2
|
dishevelled associated activator of morphogenesis 2
|
|
chr8_-_26780645
|
0.987
|
|
|
|
|
chr1_+_99502655
|
0.979
|
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr13_-_24643778
|
0.967
|
NM_152704
NM_199138
|
FAM123A
|
family with sequence similarity 123A
|
|
chr4_-_156517394
|
0.964
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr10_-_73518269
|
0.963
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr10_-_73518284
|
0.960
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr7_-_37922902
|
0.958
|
|
SFRP4
|
secreted frizzled-related protein 4
|
|
chr10_-_52315436
|
0.948
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr4_-_156517544
|
0.943
|
NM_001039580
|
MAP9
|
microtubule-associated protein 9
|
|
chr1_-_152431175
|
0.930
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr4_-_156517423
|
0.930
|
|
MAP9
|
microtubule-associated protein 9
|
|
chr11_+_108798084
|
0.912
|
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr2_+_219991342
|
0.908
|
NM_001927
|
DES
|
desmin
|
|
chr10_-_99780336
|
0.907
|
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr3_+_186563529
|
0.900
|
NM_004721
|
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13
|
|
chrX_+_117992623
|
0.893
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr2_-_213723298
|
0.871
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr1_-_203657803
|
0.867
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr10_-_73518315
|
0.856
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr4_+_54790190
|
0.846
|
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chrX_-_131918905
|
0.839
|
|
HS6ST2
|
heparan sulfate 6-O-sulfotransferase 2
|
|
chr2_-_200028977
|
0.821
|
|
SATB2
|
SATB homeobox 2
|
|
chr9_-_126573349
|
0.818
|
NM_001489
NM_033334
|
NR6A1
|
nuclear receptor subfamily 6, group A, member 1
|
|
chr12_+_52697160
|
0.816
|
|
HOXC5
HOXC6
|
homeobox C5
homeobox C6
|
|
chr7_-_100595552
|
0.815
|
NM_003378
|
VGF
|
VGF nerve growth factor inducible
|
|
chr8_-_27171852
|
0.807
|
|
STMN4
|
stathmin-like 4
|
|
chr8_-_102872354
|
0.804
|
|
NCALD
|
neurocalcin delta
|
|
chr20_+_17155680
|
0.801
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr5_-_88004891
|
0.798
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr8_-_27171814
|
0.794
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr4_+_88939725
|
0.784
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr16_+_54782802
|
0.775
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr8_-_102872372
|
0.773
|
|
NCALD
|
neurocalcin delta
|
|
chr3_+_138965844
|
0.762
|
|
SOX14
|
SRY (sex determining region Y)-box 14
|
|
chr21_+_25933459
|
0.762
|
NM_021219
|
JAM2
|
junctional adhesion molecule 2
|
|
chr15_-_70399523
|
0.760
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr19_-_56919032
|
0.760
|
NM_001523
|
HAS1
|
hyaluronan synthase 1
|
|
chr2_-_220116508
|
0.756
|
NM_024536
|
CHPF
|
chondroitin polymerizing factor
|
|
chr21_+_38550739
|
0.742
|
NM_170736
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr15_-_70399551
|
0.731
|
NM_052840
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr11_-_129803712
|
0.727
|
NM_007037
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr20_+_9442995
|
0.725
|
|
C20orf103
|
chromosome 20 open reading frame 103
|
|
chr5_+_36642213
|
0.721
|
NM_001166696
NM_004172
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_-_149955870
|
0.721
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr11_-_14950366
|
0.718
|
NM_001033952
NM_001033953
NM_001741
|
CALCA
|
calcitonin-related polypeptide alpha
|
|
chr13_-_24643620
|
0.714
|
|
FAM123A
|
family with sequence similarity 123A
|
|
chr3_-_159872960
|
0.710
|
NM_020169
|
LXN
|
latexin
|
|
chr17_+_4743645
|
0.702
|
NM_001145536
|
C17orf107
|
chromosome 17 open reading frame 107
|
|
chr1_-_199635057
|
0.691
|
|
LAD1
|
ladinin 1
|
|
chr2_-_96899326
|
0.672
|
|
SEMA4C
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C
|
|
chr17_+_12510030
|
0.670
|
|
MYOCD
|
myocardin
|
|
chr8_-_10734700
|
0.669
|
NM_017884
|
PINX1
|
PIN2/TERF1 interacting, telomerase inhibitor 1
|
|
chr8_+_26296439
|
0.668
|
NM_004331
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like
|
|
chr7_+_73080362
|
0.664
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr14_+_100362204
|
0.663
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr15_-_70399301
|
0.658
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr7_+_116907252
|
0.656
|
NM_000492
|
CFTR
|
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7)
|
|
chr17_-_23151601
|
0.655
|
NM_000625
|
NOS2
|
nitric oxide synthase 2, inducible
|
|
chr9_+_17568952
|
0.652
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr3_+_141136691
|
0.652
|
NM_022131
|
CLSTN2
|
calsyntenin 2
|
|
chrX_+_151975421
|
0.645
|
NM_013364
|
PNMA3
|
paraneoplastic antigen MA3
|
|
chr8_-_67253234
|
0.644
|
NM_000756
|
CRH
|
corticotropin releasing hormone
|
|
chr10_-_99780574
|
0.641
|
NM_018058
|
CRTAC1
|
cartilage acidic protein 1
|
|
chr12_-_7547552
|
0.637
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr16_-_2258067
|
0.629
|
|
RNPS1
|
RNA binding protein S1, serine-rich domain
|
|
chr1_+_154297588
|
0.628
|
NM_020387
|
RAB25
|
RAB25, member RAS oncogene family
|
|
chr7_+_73080387
|
0.621
|
|
ELN
|
elastin
|
|
chr17_-_23151522
|
0.616
|
|
NOS2
|
nitric oxide synthase 2, inducible
|
|
chr3_-_12561962
|
0.616
|
NM_001195279
|
LOC100129480
|
hypothetical protein LOC100129480
|
|
chr5_+_170668892
|
0.615
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr1_+_99502435
|
0.615
|
NM_001166252
NM_014839
|
LPPR4
|
lipid phosphate phosphatase-related protein type 4
|
|
chr3_-_126136261
|
0.613
|
NM_033049
|
MUC13
|
mucin 13, cell surface associated
|
|
chr21_+_38550533
|
0.612
|
NM_002243
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15
|
|
chr17_-_39340414
|
0.611
|
|
MPP2
|
membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2)
|
|
chrX_-_83644054
|
0.611
|
NM_001177478
NM_001177479
NM_144657
|
HDX
|
highly divergent homeobox
|
|
chr2_-_152664056
|
0.610
|
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr10_+_35524835
|
0.608
|
NM_182721
NM_182722
NM_182723
NM_182724
NM_182725
|
CREM
|
cAMP responsive element modulator
|
|
chr2_-_224175293
|
0.606
|
|
SCG2
|
secretogranin II
|
|
chr7_+_128099581
|
0.603
|
NM_001012454
NM_001128926
|
FAM71F2
|
family with sequence similarity 71, member F2
|
|
chr1_+_210275735
|
0.598
|
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr14_-_46882144
|
0.594
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr7_+_73080404
|
0.591
|
|
ELN
|
elastin
|
|
chr10_+_95838801
|
0.587
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr5_-_1935764
|
0.584
|
NM_016358
|
IRX4
|
iroquois homeobox 4
|
|
chr8_-_72437020
|
0.584
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr15_-_71448657
|
0.583
|
NM_005477
|
HCN4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4
|
|
chr5_-_153837975
|
0.581
|
|
HAND1
|
heart and neural crest derivatives expressed 1
|
|
chr17_-_13445942
|
0.579
|
NM_006042
|
HS3ST3A1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1
|
|
chr15_-_81412354
|
0.579
|
NM_004839
NM_199330
NM_199331
NM_199332
|
HOMER2
|
homer homolog 2 (Drosophila)
|
|
chr4_+_41395493
|
0.575
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr11_-_8572321
|
0.570
|
|
STK33
|
serine/threonine kinase 33
|
|
chr17_+_30938391
|
0.568
|
NM_001030006
NM_001282
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr14_+_71469538
|
0.568
|
NM_004296
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr19_+_50196526
|
0.567
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr14_+_20568184
|
0.563
|
NM_173846
|
TPPP2
|
tubulin polymerization-promoting protein family member 2
|
|
chr19_+_55671468
|
0.563
|
NM_175063
NM_206538
|
C19orf63
|
chromosome 19 open reading frame 63
|
|
chr17_+_26839143
|
0.555
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr4_+_113958652
|
0.554
|
NM_001127493
|
ANK2
|
ankyrin 2, neuronal
|
|
chr8_-_10734792
|
0.550
|
|
PINX1
|
PIN2/TERF1 interacting, telomerase inhibitor 1
|
|
chr4_-_44145580
|
0.549
|
NM_198353
|
KCTD8
|
potassium channel tetramerisation domain containing 8
|
|
chr16_-_3008162
|
0.547
|
NM_021195
|
CLDN6
|
claudin 6
|
|
chr20_+_17155617
|
0.541
|
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr12_-_16652202
|
0.538
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chrX_-_13745108
|
0.535
|
|
GPM6B
|
glycoprotein M6B
|
|
chr8_+_65655367
|
0.534
|
NM_152414
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr4_-_41445693
|
0.533
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chrX_-_13866451
|
0.532
|
|
GPM6B
|
glycoprotein M6B
|
|
chr2_-_135498716
|
0.530
|
NM_001018046
NM_025052
|
YSK4
|
YSK4 Sps1/Ste20-related kinase homolog (S. cerevisiae)
|
|
chr2_-_133895795
|
0.530
|
|
NCKAP5
|
NCK-associated protein 5
|
|
chr14_-_79748273
|
0.527
|
NM_000793
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr17_+_41945391
|
0.527
|
NM_001006607
|
LRRC37A2
LRRC37A
|
leucine rich repeat containing 37, member A2
leucine rich repeat containing 37A
|
|
chrX_-_47364173
|
0.526
|
NM_006950
NM_133499
|
SYN1
|
synapsin I
|
|
chr10_+_68355797
|
0.522
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr7_-_121731721
|
0.522
|
NM_001024613
NM_001160264
|
FEZF1
|
FEZ family zinc finger 1
|
|
chr14_+_100362236
|
0.522
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr11_-_129803515
|
0.518
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr11_-_35397675
|
0.517
|
NM_004171
|
SLC1A2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr16_+_54782751
|
0.514
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr8_-_42742729
|
0.510
|
NM_004198
|
CHRNA6
|
cholinergic receptor, nicotinic, alpha 6
|
|
chr7_+_73080422
|
0.507
|
|
ELN
|
elastin
|
|
chr8_+_134272463
|
0.506
|
NM_003882
NM_080838
|
WISP1
|
WNT1 inducible signaling pathway protein 1
|
|
chr12_-_7935010
|
0.504
|
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr17_+_38306339
|
0.500
|
NM_000151
|
G6PC
|
glucose-6-phosphatase, catalytic subunit
|
|
chr1_-_210275402
|
0.500
|
NM_015434
|
INTS7
|
integrator complex subunit 7
|
|
chr10_+_50557689
|
0.498
|
NM_001042427
NM_182554
|
C10orf53
|
chromosome 10 open reading frame 53
|
|
chr18_+_13208777
|
0.494
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr17_-_75385371
|
0.494
|
NM_020649
|
CBX8
|
chromobox homolog 8
|
|
chr9_+_126460511
|
0.492
|
|
|
|
|
chr1_+_177261678
|
0.492
|
NM_014864
|
FAM20B
|
family with sequence similarity 20, member B
|
|
chr5_+_118719591
|
0.489
|
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8
|
|
chrX_-_13745134
|
0.486
|
|
GPM6B
|
glycoprotein M6B
|
|
chr15_-_29950070
|
0.485
|
|
OTUD7A
|
OTU domain containing 7A
|
|
chr11_+_117735502
|
0.484
|
NM_004788
|
UBE4A
|
ubiquitination factor E4A (UFD2 homolog, yeast)
|
|
chr22_+_40286957
|
0.482
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr1_+_210275541
|
0.480
|
NM_016448
|
DTL
|
denticleless homolog (Drosophila)
|
|
chr20_-_13919256
|
0.479
|
NM_025229
|
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr14_+_92721048
|
0.477
|
NM_015676
NM_001098621
|
C14orf109
|
chromosome 14 open reading frame 109
|
|
chr1_-_7835681
|
0.476
|
NM_006786
|
UTS2
|
urotensin 2
|
|
chr5_+_126140213
|
0.476
|
NM_005573
|
LMNB1
|
lamin B1
|
|
chr19_+_60487345
|
0.475
|
NM_032430
|
BRSK1
|
BR serine/threonine kinase 1
|
|
chr1_+_7766966
|
0.474
|
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr12_+_70120028
|
0.473
|
NM_003667
|
LGR5
|
leucine-rich repeat containing G protein-coupled receptor 5
|
|
chr15_-_70399181
|
0.468
|
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr15_-_21444085
|
0.467
|
NM_019066
|
MAGEL2
|
MAGE-like 2
|
|
chr11_+_24475091
|
0.466
|
NM_001009909
|
LUZP2
|
leucine zipper protein 2
|
|
chrX_-_47364039
|
0.460
|
|
|
|
|
chr8_-_26780665
|
0.460
|
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chrX_+_50044279
|
0.459
|
NM_033031
NM_033670
|
CCNB3
|
cyclin B3
|
|
chrX_-_13745234
|
0.459
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr6_+_97117144
|
0.459
|
NM_001170807
NM_020482
|
FHL5
|
four and a half LIM domains 5
|
|
chr15_+_36776090
|
0.459
|
NM_207444
|
C15orf53
|
chromosome 15 open reading frame 53
|