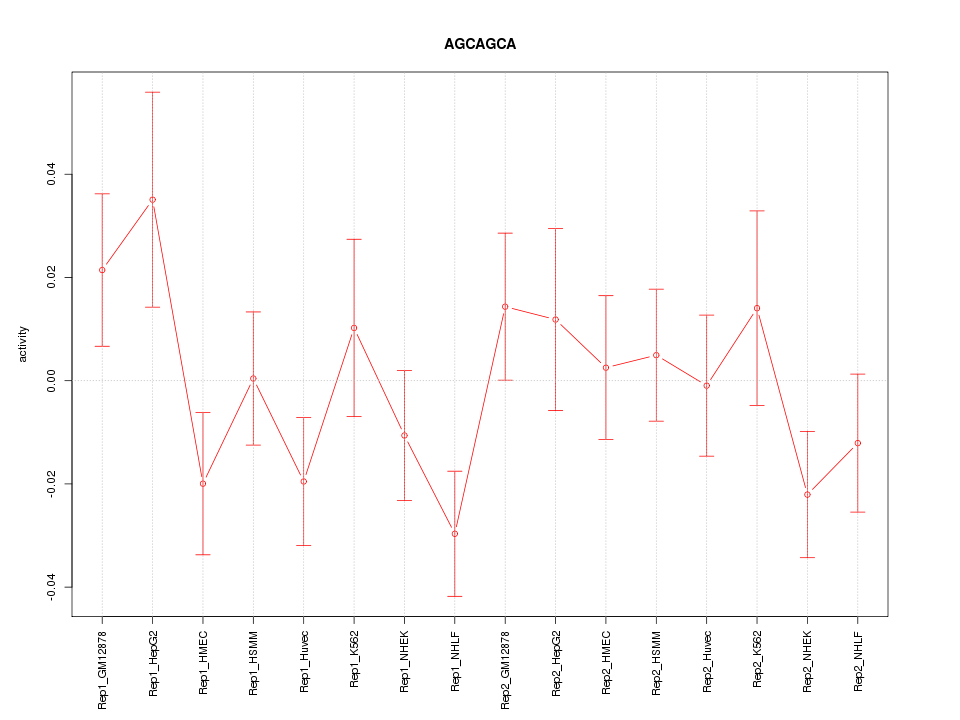
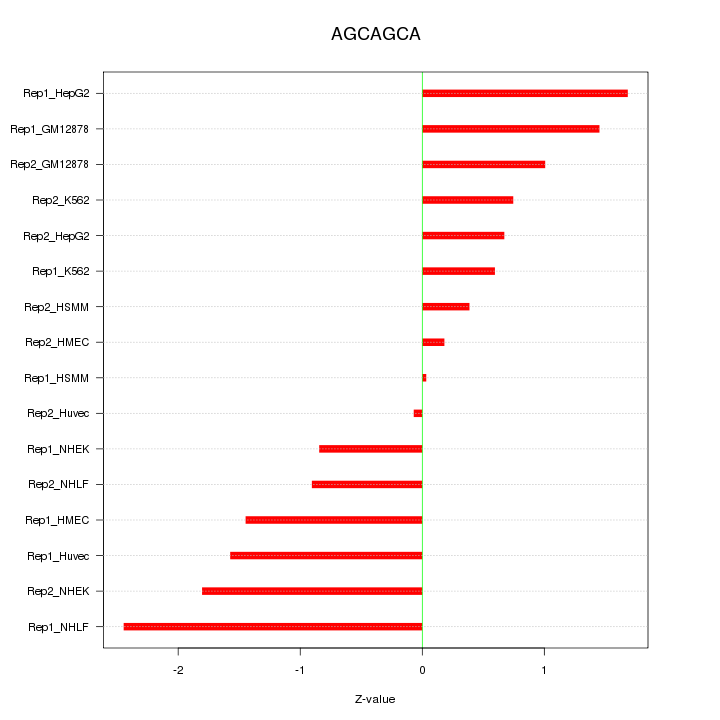
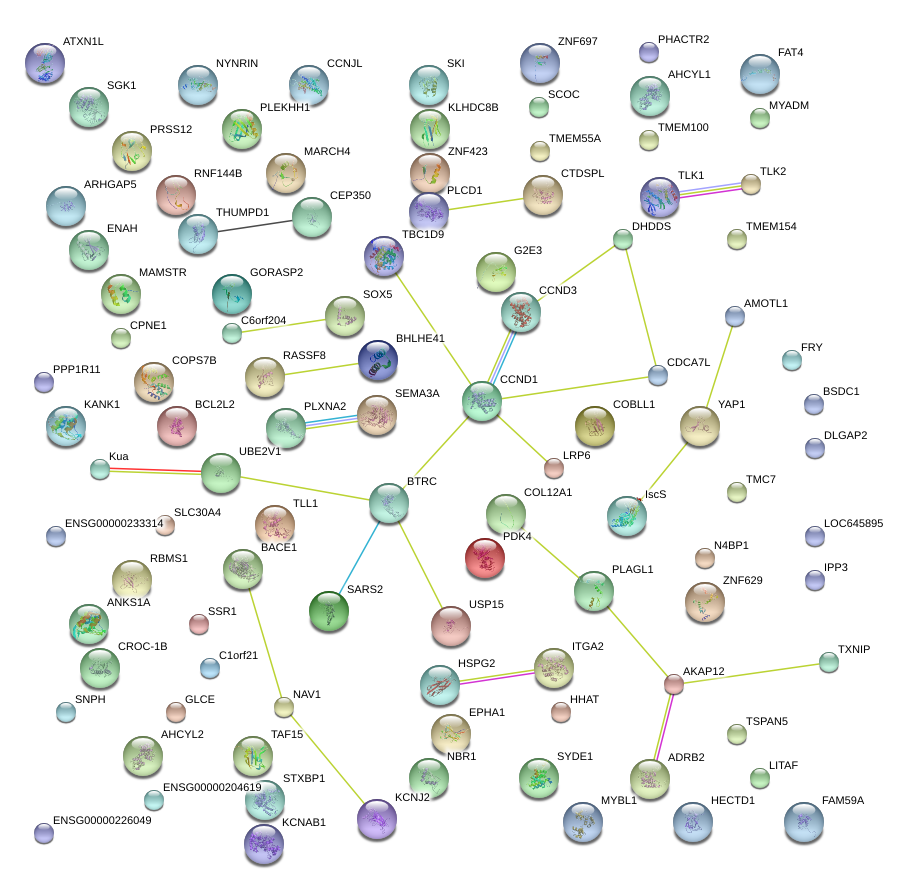
|
chr1_-_206484258
|
1.951
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr4_-_99798563
|
1.764
|
NM_005723
|
TSPAN5
|
tetraspanin 5
|
|
chr6_+_18495559
|
1.560
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr1_-_223907283
|
1.531
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr12_-_24606616
|
1.497
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr2_-_216944911
|
1.464
|
NM_020814
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4
|
|
chr2_-_161058550
|
1.449
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr9_+_696744
|
1.418
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr14_+_22845851
|
1.415
|
NM_004050
|
BCL2L2-PABPN1
BCL2L2
|
BCL2L2-PABPN1 read-through transcript
BCL2-like 2
|
|
chr14_+_23937766
|
1.398
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr11_+_94141151
|
1.393
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr16_-_30705707
|
1.358
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr6_+_151688327
|
1.295
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr3_+_37878647
|
1.268
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr6_-_134680888
|
1.251
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_+_30098079
|
1.213
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr18_-_28304392
|
1.148
|
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr14_+_31616196
|
1.139
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr12_-_26169074
|
1.114
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr17_+_65677255
|
1.106
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr6_-_75972286
|
1.102
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr12_-_12310969
|
1.099
|
NM_002336
|
LRP6
|
low density lipoprotein receptor-related protein 6
|
|
chr11_+_101485946
|
1.088
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr5_+_52320660
|
1.081
|
NM_002203
|
ITGA2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor)
|
|
chr1_+_199975563
|
1.046
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr15_-_43602293
|
1.040
|
NM_013309
|
SLC30A4
|
solute carrier family 30 (zinc transporter), member 4
|
|
chr8_-_92122219
|
1.029
|
NM_018710
|
TMEM55A
|
transmembrane protein 55A
|
|
chr8_-_67687980
|
1.025
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
hypothetical LOC645895
|
|
chr7_-_83662152
|
1.025
|
NM_006080
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr4_+_141397889
|
1.024
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr15_+_67239853
|
1.007
|
NM_015554
|
GLCE
|
glucuronic acid epimerase
|
|
chr4_-_119493322
|
0.979
|
NM_003619
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin)
|
|
chr20_-_48163071
|
0.977
|
NM_001032288
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr6_-_119137923
|
0.960
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr17_-_51155073
|
0.952
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr4_-_153820535
|
0.951
|
NM_152680
|
TMEM154
|
transmembrane protein 154
|
|
chr2_-_171725604
|
0.914
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr17_-_51164443
|
0.911
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr14_-_30746431
|
0.906
|
NM_015382
|
HECTD1
|
HECT domain containing 1
|
|
chr17_+_38576707
|
0.896
|
NM_005899
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr12_+_60940404
|
0.894
|
NM_006313
|
USP15
|
ubiquitin specific peptidase 15
|
|
chr19_+_15079141
|
0.888
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr20_+_1194959
|
0.863
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr6_-_42017337
|
0.860
|
NM_001136125
NM_001760
|
CCND3
|
cyclin D3
|
|
chr7_-_95063738
|
0.836
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr16_+_18903009
|
0.829
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr14_+_67069668
|
0.829
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr20_-_33750457
|
0.828
|
NM_001198989
NM_021100
|
NFS1
|
NFS1 nitrogen fixation 1 homolog (S. cerevisiae)
|
|
chr7_-_21951946
|
0.821
|
NM_001127370
NM_001127371
NM_018719
|
CDCA7L
|
cell division cycle associated 7-like
|
|
chr6_+_34964999
|
0.812
|
NM_015245
|
ANKS1A
|
ankyrin repeat and sterile alpha motif domain containing 1A
|
|
chr4_+_126457016
|
0.811
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr2_+_232359303
|
0.809
|
NM_022730
|
COPS7B
|
COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis)
|
|
chr11_-_116692128
|
0.804
|
NM_012104
NM_138971
NM_138972
NM_138973
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr19_-_53914768
|
0.787
|
NM_001130915
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr19_-_44158130
|
0.783
|
NM_024907
|
FBXO17
|
F-box protein 17
|
|
chr4_+_167013859
|
0.781
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr13_+_31503436
|
0.780
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr12_+_26003218
|
0.774
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_-_119991912
|
0.771
|
NM_001080470
|
ZNF697
|
zinc finger protein 697
|
|
chr6_+_144040751
|
0.762
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr12_+_26017954
|
0.753
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr6_+_30142910
|
0.751
|
NM_021959
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11
|
|
chr11_+_101488380
|
0.750
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr11_+_69165018
|
0.748
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr4_-_141896920
|
0.744
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr16_-_20660618
|
0.744
|
NM_017736
|
THUMPD1
|
THUMP domain containing 1
|
|
chr12_+_26096757
|
0.738
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr1_+_144149794
|
0.737
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr3_-_38046034
|
0.736
|
NM_006225
|
PLCD1
|
phospholipase C, delta 1
|
|
chr9_+_129414287
|
0.730
|
NM_001032221
NM_003165
|
STXBP1
|
syntaxin binding protein 1
|
|
chr16_-_48418418
|
0.730
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr1_-_22136263
|
0.728
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr7_+_128651976
|
0.720
|
NM_001130720
NM_015328
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr10_+_103103797
|
0.708
|
NM_003939
NM_033637
|
BTRC
|
beta-transducin repeat containing
|
|
chr6_-_7258351
|
0.686
|
NM_003144
|
SSR1
|
signal sequence receptor, alpha
|
|
chr3_+_49183983
|
0.685
|
NM_173546
|
KLHDC8B
|
kelch domain containing 8B
|
|
chr1_-_32632601
|
0.680
|
NM_001143888
NM_001143889
NM_001143890
NM_018045
|
BSDC1
|
BSD domain containing 1
|
|
chr16_+_70437394
|
0.673
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr5_-_159672060
|
0.667
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr8_+_1436938
|
0.660
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr16_-_11587647
|
0.654
|
NM_004862
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr6_-_144371035
|
0.640
|
NM_001080952
NM_001080953
NM_001080954
NM_001080956
NM_002656
NM_006718
|
PLAGL1
HYMAI
|
pleiomorphic adenoma gene-like 1
hydatidiform mole associated and imprinted (non-protein coding)
|
|
chr3_+_157343444
|
0.637
|
NM_003471
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr1_+_178190493
|
0.637
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr1_+_26631359
|
0.636
|
NM_024887
NM_205861
|
DHDDS
|
dehydrodolichyl diphosphate synthase
|
|
chr2_-_165406173
|
0.635
|
NM_014900
|
COBLL1
|
COBL-like 1
|
|
chr16_-_47201588
|
0.632
|
NM_153029
|
N4BP1
|
NEDD4 binding protein 1
|
|
chr5_+_148186348
|
0.626
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr2_+_171493870
|
0.625
|
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr7_-_142816019
|
0.625
|
NM_005232
|
EPHA1
|
EPH receptor A1
|
|
chr19_+_59061422
|
0.619
|
NM_001020820
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr1_+_182622751
|
0.617
|
NM_030806
|
C1orf21
|
chromosome 1 open reading frame 21
|
|
chr1_+_2149993
|
0.615
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr17_+_31160550
|
0.610
|
NM_003487
NM_139215
|
TAF15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa
|
|
chr9_+_4669557
|
0.607
|
NM_017913
|
CDC37L1
|
cell division cycle 37 homolog (S. cerevisiae)-like 1
|
|
chr1_-_181540631
|
0.606
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr7_-_138316393
|
0.604
|
NM_001164665
NM_020910
|
KIAA1549
|
KIAA1549
|
|
chr8_+_79740813
|
0.602
|
NM_016010
|
FAM164A
|
family with sequence similarity 164, member A
|
|
chr18_-_63334902
|
0.601
|
NM_032160
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr16_+_18902756
|
0.600
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr1_-_120413798
|
0.600
|
NM_024408
|
NOTCH2
|
notch 2
|
|
chr11_+_86426518
|
0.599
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr1_-_85439315
|
0.595
|
NM_032184
|
SYDE2
|
synapse defective 1, Rho GTPase, homolog 2 (C. elegans)
|
|
chr6_+_143971009
|
0.594
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr9_+_117955890
|
0.593
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr2_+_206255376
|
0.591
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr15_+_89212888
|
0.591
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr9_+_131974506
|
0.589
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr11_+_117812282
|
0.588
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr20_-_16501911
|
0.588
|
NM_024704
|
KIF16B
|
kinesin family member 16B
|
|
chr6_+_11202251
|
0.585
|
NM_001135575
|
LOC221710
|
hypothetical protein LOC221710
|
|
chr1_+_46441592
|
0.584
|
NM_001013615
|
C1orf190
|
chromosome 1 open reading frame 190
|
|
chr8_+_22513043
|
0.582
|
NM_001013842
NM_001198827
NM_173686
|
C8orf58
|
chromosome 8 open reading frame 58
|
|
chr17_+_11864841
|
0.580
|
NM_003010
|
MAP2K4
|
mitogen-activated protein kinase kinase 4
|
|
chr1_+_165456572
|
0.576
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr17_-_34860840
|
0.574
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr1_-_115014151
|
0.573
|
NM_198459
|
DENND2C
|
DENN/MADD domain containing 2C
|
|
chr14_-_87806913
|
0.571
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr20_+_56397568
|
0.571
|
NM_001195677
NM_004738
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C
|
|
chr2_+_203901160
|
0.566
|
NM_005759
|
ABI2
|
abl-interactor 2
|
|
chr1_+_169721260
|
0.565
|
NM_015172
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr17_-_46140015
|
0.558
|
NM_052855
|
ANKRD40
|
ankyrin repeat domain 40
|
|
chr12_-_55316350
|
0.558
|
NM_013449
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A
|
|
chr10_+_124135574
|
0.558
|
NM_001195608
|
PLEKHA1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1
|
|
chr3_+_43303005
|
0.557
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr16_+_84202515
|
0.555
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr2_-_121759237
|
0.553
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr2_-_73193581
|
0.552
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr3_-_121652183
|
0.547
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr12_-_105057940
|
0.546
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr5_-_54866126
|
0.546
|
NM_003711
NM_176895
|
RNF138P1
PPAP2A
|
ring finger protein 138 pseudogene 1
phosphatidic acid phosphatase type 2A
|
|
chr18_+_7557313
|
0.544
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chrX_-_128616594
|
0.539
|
NM_017413
|
APLN
|
apelin
|
|
chr6_-_40010118
|
0.530
|
NM_001075098
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr12_+_55201854
|
0.528
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr6_-_44333002
|
0.527
|
NM_178148
|
SLC35B2
|
solute carrier family 35, member B2
|
|
chr16_+_10945815
|
0.526
|
NM_015226
|
CLEC16A
|
C-type lectin domain family 16, member A
|
|
chr2_-_39201066
|
0.524
|
NM_005633
|
SOS1
|
son of sevenless homolog 1 (Drosophila)
|
|
chr8_-_38444398
|
0.521
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr6_+_139136349
|
0.521
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr3_-_48569213
|
0.521
|
NM_004567
|
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
|
|
chr8_+_41554823
|
0.518
|
NM_178819
|
AGPAT6
|
1-acylglycerol-3-phosphate O-acyltransferase 6 (lysophosphatidic acid acyltransferase, zeta)
|
|
chr11_-_34336102
|
0.518
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr15_+_36332258
|
0.516
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr20_-_48165886
|
0.516
|
NM_022442
NM_021988
NM_199144
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr2_+_148319033
|
0.514
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr22_+_27609889
|
0.514
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr14_+_23905984
|
0.512
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr1_+_23990228
|
0.509
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr3_-_125085834
|
0.508
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr17_-_44794325
|
0.507
|
NM_001145365
NM_014897
|
ZNF652
|
zinc finger protein 652
|
|
chr14_-_60260211
|
0.506
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr16_+_51646292
|
0.500
|
NM_025134
|
CHD9
|
chromodomain helicase DNA binding protein 9
|
|
chr20_-_13567529
|
0.499
|
NM_017714
|
TASP1
|
taspase, threonine aspartase, 1
|
|
chr1_+_24158887
|
0.498
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr14_-_104515738
|
0.498
|
NM_138420
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr22_-_34754343
|
0.490
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr1_+_165564904
|
0.490
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_+_151602787
|
0.489
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr14_-_38971370
|
0.488
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr15_+_73728402
|
0.488
|
NM_153271
|
SNX33
|
sorting nexin 33
|
|
chr1_+_78243181
|
0.486
|
NM_007034
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4
|
|
chr2_-_142605739
|
0.486
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr5_+_67558217
|
0.484
|
NM_181523
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr8_+_32623792
|
0.484
|
NM_013959
|
NRG1
|
neuregulin 1
|
|
chr7_-_105304159
|
0.482
|
NM_020725
NM_152749
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr16_-_71639670
|
0.482
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr14_+_36736867
|
0.477
|
NM_001195296
NM_001195297
NM_138731
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr7_-_27125738
|
0.474
|
NM_030661
|
HOXA3
|
homeobox A3
|
|
chr6_+_20510019
|
0.472
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr2_-_128358903
|
0.470
|
NM_031445
|
AMMECR1L
|
AMME chromosomal region gene 1-like
|
|
chr6_-_138470160
|
0.469
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr8_-_93181091
|
0.468
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_+_87424074
|
0.467
|
NM_007013
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr5_+_67622242
|
0.465
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_-_2696152
|
0.464
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr14_-_22891865
|
0.463
|
NM_016609
|
SLC22A17
|
solute carrier family 22, member 17
|
|
chr14_-_52487558
|
0.462
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr20_+_34206070
|
0.460
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr17_+_4683280
|
0.458
|
NM_001024937
NM_015716
NM_153827
NM_170663
|
MINK1
|
misshapen-like kinase 1
|
|
chr3_-_51976428
|
0.453
|
NM_020418
NM_001174100
NM_033008
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr2_-_166518500
|
0.451
|
NM_024753
|
TTC21B
|
tetratricopeptide repeat domain 21B
|
|
chr1_+_42896634
|
0.450
|
NM_006347
|
PPIH
|
peptidylprolyl isomerase H (cyclophilin H)
|
|
chr1_+_33494745
|
0.447
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr15_-_58671929
|
0.445
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr17_-_34811377
|
0.444
|
NM_001184906
NM_032875
|
FBXL20
|
F-box and leucine-rich repeat protein 20
|
|
chr4_+_154345039
|
0.442
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr3_-_12680626
|
0.442
|
NM_002880
|
RAF1
|
v-raf-1 murine leukemia viral oncogene homolog 1
|
|
chr5_+_172000793
|
0.441
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr9_-_98421894
|
0.441
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr15_+_41597097
|
0.439
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr13_-_46368968
|
0.439
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr12_+_3056781
|
0.437
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr22_-_36245155
|
0.435
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr12_-_55686466
|
0.434
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr11_-_117528744
|
0.434
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr4_+_123967265
|
0.433
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr17_-_53950191
|
0.432
|
NM_004687
|
MTMR4
|
myotubularin related protein 4
|
|
chr15_+_39032927
|
0.431
|
NM_001142776
NM_024111
|
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli)
|
|
chr20_+_34163671
|
0.430
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|