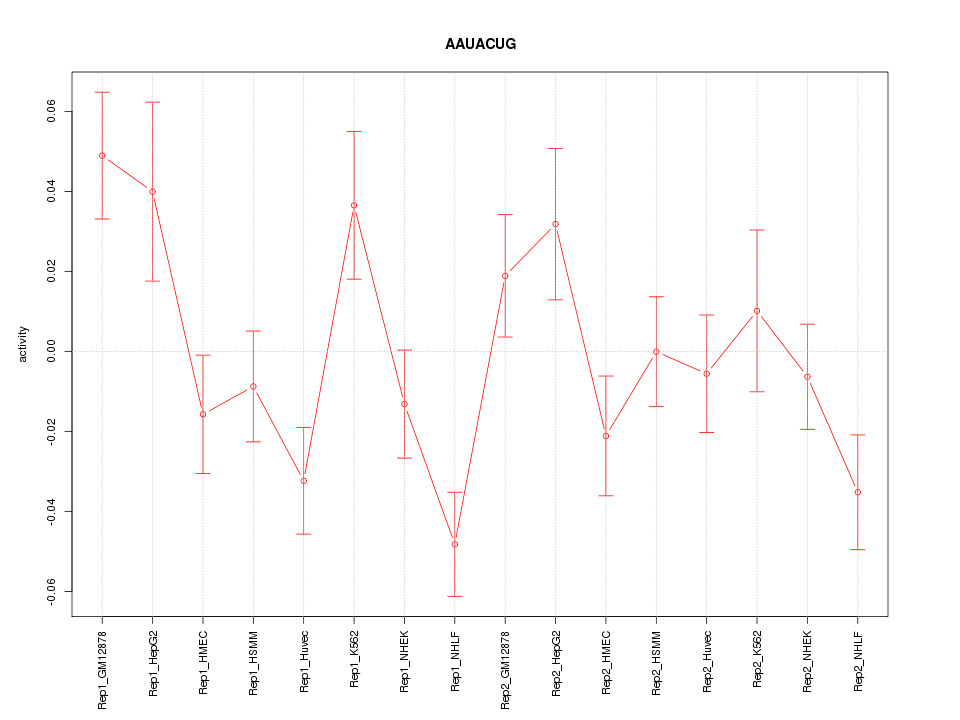
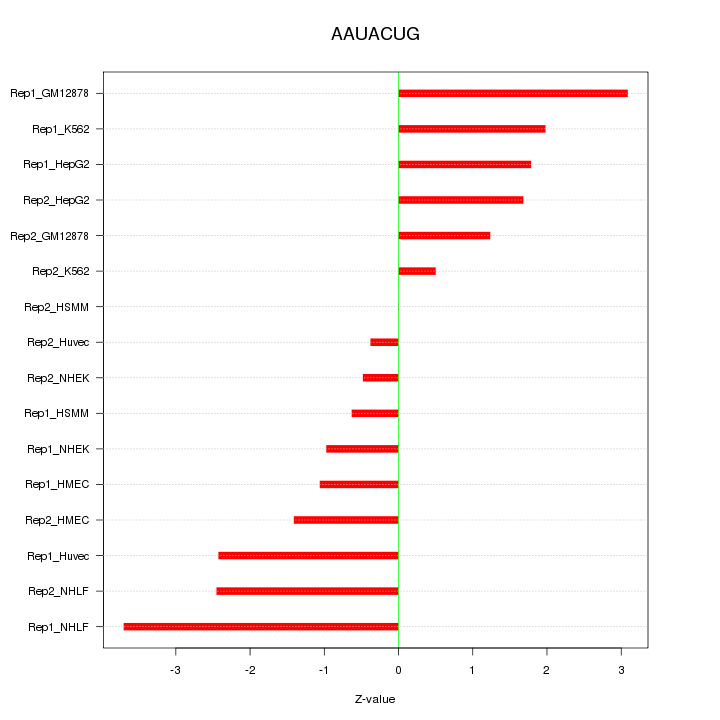
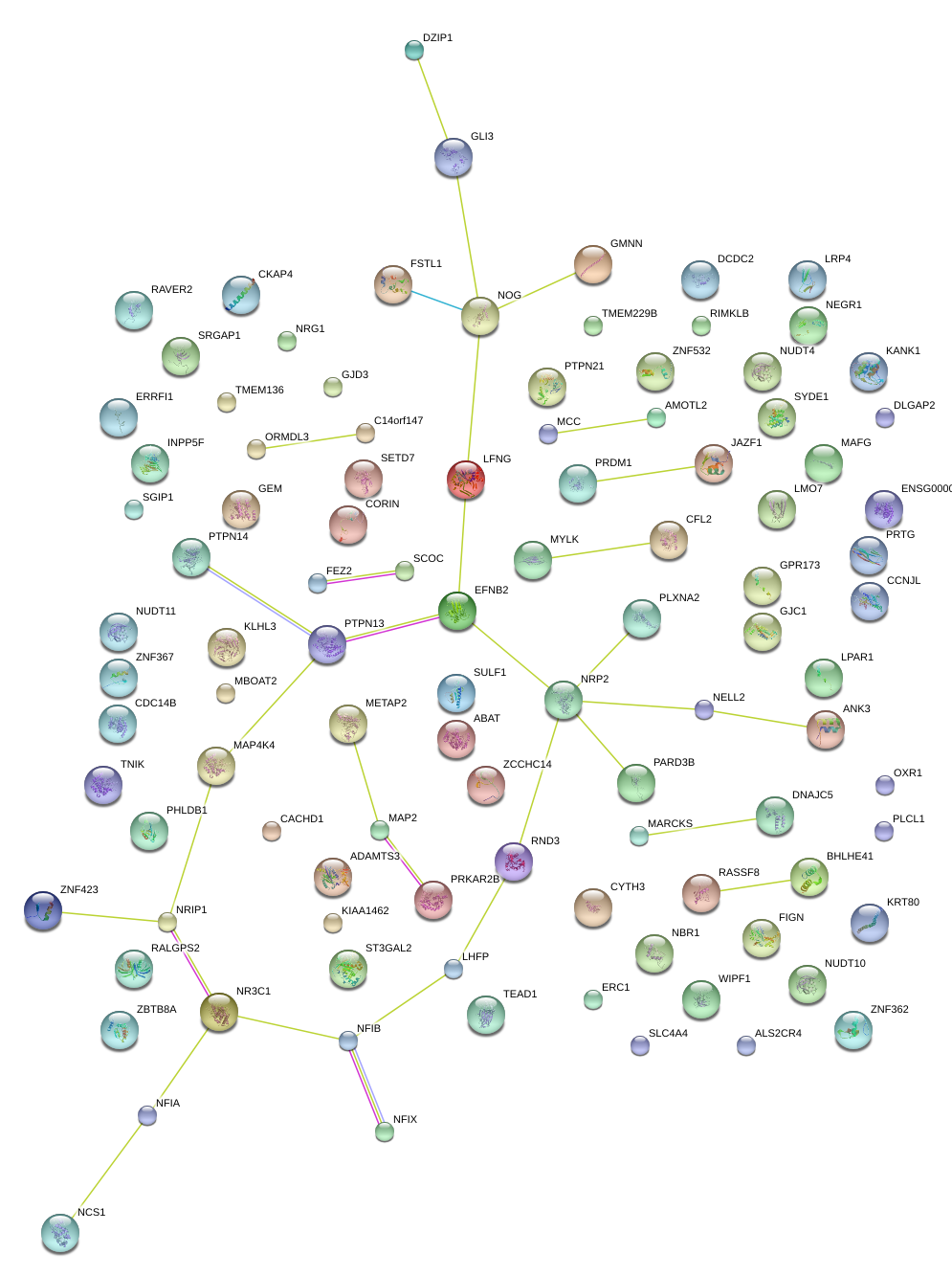
|
chr17_-_40263704
|
3.488
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr17_-_40263124
|
3.165
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr7_-_28186867
|
3.034
|
NM_175061
|
JAZF1
|
JAZF zinc finger 1
|
|
chr1_-_206484258
|
2.926
|
NM_025179
|
PLXNA2
|
plexin A2
|
|
chr17_+_52026058
|
2.746
|
NM_005450
|
NOG
|
noggin
|
|
chr2_-_151052391
|
2.410
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr5_-_112658456
|
2.333
|
NM_002387
|
MCC
|
mutated in colorectal cancers
|
|
chr22_-_34754343
|
2.318
|
NM_001082578
NM_001082579
|
RBFOX2
|
RNA binding protein, fox-1 homolog (C. elegans) 2
|
|
chr9_+_696744
|
2.280
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr11_-_46896484
|
2.145
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chr5_-_112852425
|
2.130
|
NM_001085377
|
MCC
|
mutated in colorectal cancers
|
|
chr3_-_135575925
|
2.087
|
NM_016201
|
AMOTL2
|
angiomotin like 2
|
|
chr19_+_15079141
|
2.027
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr3_-_121652183
|
2.004
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr1_-_8008979
|
1.936
|
NM_018948
|
ERRFI1
|
ERBB receptor feedback inhibitor 1
|
|
chr2_+_101680594
|
1.816
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr11_+_119701047
|
1.807
|
NM_001198670
NM_001198671
NM_001198672
NM_001198673
NM_001198674
NM_001198675
NM_174926
|
TMEM136
|
transmembrane protein 136
|
|
chr1_+_176960896
|
1.765
|
NM_152663
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr7_+_2524022
|
1.714
|
NM_002304
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr1_+_61320213
|
1.702
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr2_-_164300758
|
1.696
|
NM_018086
|
FIGN
|
fidgetin
|
|
chr1_+_66772393
|
1.534
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr12_+_8741746
|
1.528
|
NM_020734
|
RIMKLB
|
ribosomal modification protein rimK-like family member B
|
|
chr4_-_73653331
|
1.527
|
NM_014243
|
ADAMTS3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3
|
|
chr3_-_125085834
|
1.514
|
NM_053025
NM_053026
NM_053027
NM_053028
|
MYLK
|
myosin light chain kinase
|
|
chr12_+_62524778
|
1.498
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr13_+_75232797
|
1.466
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chr14_-_34001164
|
1.450
|
NM_138288
|
C14orf147
|
chromosome 14 open reading frame 147
|
|
chr13_-_95094920
|
1.441
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|
|
chr10_-_61819493
|
1.441
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr12_+_92295738
|
1.433
|
NM_019094
NM_199040
|
NUDT4
NUDT4P1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
nudix (nucleoside diphosphate linked moiety X)-type motif 4 pseudogene 1
|
|
chr1_+_61319976
|
1.431
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr12_+_26003218
|
1.424
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr14_-_88090641
|
1.376
|
NM_007039
|
PTPN21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr17_+_38576707
|
1.364
|
NM_005899
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr2_-_36678657
|
1.363
|
NM_001042548
NM_005102
|
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II)
|
|
chr9_+_131974506
|
1.361
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr12_+_26017954
|
1.346
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr2_+_205118710
|
1.340
|
NM_057177
NM_152526
NM_205863
|
PARD3B
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chrX_+_53095230
|
1.297
|
NM_018969
|
GPR173
|
G protein-coupled receptor 173
|
|
chr2_+_198377561
|
1.293
|
NM_006226
|
PLCL1
|
phospholipase C-like 1
|
|
chr2_-_9061193
|
1.289
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr2_-_175255717
|
1.288
|
NM_001077269
|
WIPF1
|
WAS/WASL interacting protein family, member 1
|
|
chr10_-_30388439
|
1.265
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr2_+_209996913
|
1.233
|
NM_001039538
|
MAP2
|
microtubule-associated protein 2
|
|
chr5_-_137099602
|
1.223
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr1_+_64709063
|
1.222
|
NM_020925
|
CACHD1
|
cache domain containing 1
|
|
chr12_-_105165805
|
1.213
|
NM_006825
|
CKAP4
|
cytoskeleton-associated protein 4
|
|
chr16_-_69030421
|
1.211
|
NM_006927
|
ST3GAL2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2
|
|
chr9_-_112840064
|
1.205
|
NM_001401
NM_057159
|
LPAR1
|
lysophosphatidic acid receptor 1
|
|
chr16_-_48418418
|
1.184
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr7_-_6278662
|
1.178
|
NM_004227
|
CYTH3
|
cytohesin 3
|
|
chr12_-_26169074
|
1.172
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr12_+_26096757
|
1.169
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr15_-_53822468
|
1.162
|
NM_173814
|
PRTG
|
protogenin
|
|
chr1_+_32777358
|
1.158
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr17_-_77474597
|
1.154
|
NM_032711
|
MAFG
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian)
|
|
chr11_+_117983515
|
1.142
|
NM_001144758
NM_001144759
|
PHLDB1
|
pleckstrin homology-like domain, family B, member 1
|
|
chr5_-_142795269
|
1.134
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr1_-_212791597
|
1.127
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr7_+_106472243
|
1.112
|
NM_002736
|
PRKAR2B
|
protein kinase, cAMP-dependent, regulatory, type II, beta
|
|
chr1_-_72520735
|
1.104
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr1_+_61315516
|
1.103
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr8_-_95343716
|
1.099
|
NM_005261
NM_181702
|
GEM
|
GTP binding protein overexpressed in skeletal muscle
|
|
chr17_-_35337288
|
1.088
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chr2_-_202216451
|
1.087
|
NM_001044385
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chr7_-_42243142
|
1.080
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr4_-_140696740
|
1.074
|
NM_030648
|
SETD7
|
SET domain containing (lysine methyltransferase) 7
|
|
chr14_-_67051773
|
1.060
|
NM_182526
|
TMEM229B
|
transmembrane protein 229B
|
|
chr12_+_970613
|
1.059
|
NM_178039
NM_178040
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1
|
|
chr13_-_39075355
|
1.057
|
NM_005780
|
LHFP
|
lipoma HMGIC fusion partner
|
|
chr16_-_86082818
|
1.051
|
NM_015144
|
ZCCHC14
|
zinc finger, CCHC domain containing 14
|
|
chr2_+_206255376
|
1.045
|
NM_003872
NM_018534
NM_201264
NM_201266
NM_201267
NM_201279
|
NRP2
|
neuropilin 2
|
|
chr1_+_64983312
|
1.045
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr6_+_106640881
|
1.040
|
NM_001198
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr5_-_159672060
|
1.037
|
NM_024565
|
CCNJL
|
cyclin J-like
|
|
chr9_-_98421894
|
1.036
|
NM_003671
NM_033331
|
CDC14B
|
CDC14 cell division cycle 14 homolog B (S. cerevisiae)
|
|
chr6_-_24491498
|
1.012
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr6_+_114285209
|
0.960
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr8_+_70541412
|
0.953
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr2_+_210152609
|
0.947
|
NM_002374
NM_031845
NM_031847
|
MAP2
|
microtubule-associated protein 2
|
|
chr13_-_105985313
|
0.946
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr4_+_72271218
|
0.942
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr8_+_1436938
|
0.939
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr12_-_50871953
|
0.936
|
NM_001081492
NM_182507
|
KRT80
|
keratin 80
|
|
chr16_+_8675904
|
0.929
|
NM_020686
|
ABAT
|
4-aminobutyrate aminotransferase
|
|
chr14_-_34253473
|
0.928
|
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr4_+_87734489
|
0.927
|
NM_006264
NM_080683
NM_080684
NM_080685
|
PTPN13
|
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase)
|
|
chr4_+_141397889
|
0.920
|
NM_032547
|
SCOC
|
short coiled-coil protein
|
|
chr20_+_61996954
|
0.917
|
NM_025219
|
DNAJC5
|
DnaJ (Hsp40) homolog, subfamily C, member 5
|
|
chr8_+_32525269
|
0.915
|
NM_001160002
NM_001160004
NM_001160005
NM_001160007
NM_001160008
NM_004495
NM_013956
NM_013957
NM_013958
NM_013960
NM_013964
|
NRG1
|
neuregulin 1
|
|
chr1_+_33494745
|
0.914
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr11_+_12652427
|
0.897
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr8_+_107351519
|
0.892
|
NM_001198533
NM_018002
|
LOC346887
OXR1
|
similar to solute carrier family 16 (monocarboxylic acid transporters), member 14
oxidation resistance 1
|
|
chr18_+_54681040
|
0.891
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr21_-_15358996
|
0.890
|
NM_003489
|
NRIP1
|
nuclear receptor interacting protein 1
|
|
chr8_+_12914143
|
0.879
|
NM_001099677
|
KIAA1456
|
KIAA1456
|
|
chr17_-_57497380
|
0.870
|
NM_005121
|
MED13
|
mediator complex subunit 13
|
|
chr16_+_84202515
|
0.867
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr6_+_106653429
|
0.866
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr3_-_38666122
|
0.864
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr15_-_54322767
|
0.857
|
NM_022841
|
RFX7
|
regulatory factor X, 7
|
|
chr2_-_166940710
|
0.829
|
NM_002977
|
SCN9A
|
sodium channel, voltage-gated, type IX, alpha subunit
|
|
chr12_-_15833617
|
0.829
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr2_-_144994349
|
0.828
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr17_-_34860840
|
0.821
|
NM_004774
|
MED1
|
mediator complex subunit 1
|
|
chr9_+_35479989
|
0.810
|
NM_014806
|
RUSC2
|
RUN and SH3 domain containing 2
|
|
chr4_-_101330629
|
0.808
|
NM_145244
|
DDIT4L
|
DNA-damage-inducible transcript 4-like
|
|
chr6_-_17814596
|
0.804
|
NM_005124
|
NUP153
|
nucleoporin 153kDa
|
|
chr9_-_138560058
|
0.803
|
NM_017617
|
NOTCH1
|
notch 1
|
|
chr20_+_45564007
|
0.802
|
NM_001174087
NM_001174088
NM_006534
NM_181659
|
NCOA3
|
nuclear receptor coactivator 3
|
|
chr7_+_2525922
|
0.796
|
NM_001040167
NM_001040168
|
LFNG
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase
|
|
chr6_-_119137923
|
0.796
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr16_-_22293097
|
0.795
|
NM_001802
|
CDR2
|
cerebellar degeneration-related protein 2, 62kDa
|
|
chr6_-_10527772
|
0.793
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr16_+_88515917
|
0.791
|
NM_001197181
|
TUBB3
|
tubulin, beta 3
|
|
chr20_+_62266270
|
0.790
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr17_-_74990157
|
0.784
|
NM_001082575
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr1_+_181258952
|
0.770
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr14_-_38971370
|
0.768
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr14_+_76297885
|
0.763
|
NM_014909
|
VASH1
|
vasohibin 1
|
|
chr14_+_51188289
|
0.756
|
NM_152330
|
FRMD6
|
FERM domain containing 6
|
|
chr7_-_139123906
|
0.748
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr3_-_126576693
|
0.746
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr19_-_52308848
|
0.746
|
NM_015168
|
ZC3H4
|
zinc finger CCCH-type containing 4
|
|
chr16_+_88517187
|
0.745
|
NM_006086
|
TUBB3
|
tubulin, beta 3
|
|
chr12_+_118590039
|
0.741
|
NM_006253
|
PRKAB1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr6_+_20510019
|
0.740
|
NM_001949
|
E2F3
|
E2F transcription factor 3
|
|
chr10_-_105604942
|
0.734
|
NM_014631
|
SH3PXD2A
|
SH3 and PX domains 2A
|
|
chr1_+_199519202
|
0.724
|
NM_000299
NM_001005337
|
PKP1
|
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome)
|
|
chr14_+_51025604
|
0.717
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr18_-_19420438
|
0.711
|
NM_000271
|
NPC1
|
Niemann-Pick disease, type C1
|
|
chr3_-_126257425
|
0.710
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr20_-_23350038
|
0.703
|
NM_022080
|
NAPB
|
N-ethylmaleimide-sensitive factor attachment protein, beta
|
|
chr17_-_74433048
|
0.700
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr14_-_34252695
|
0.699
|
NM_021914
|
CFL2
|
cofilin 2 (muscle)
|
|
chr17_+_24742068
|
0.696
|
NM_020791
|
TAOK1
|
TAO kinase 1
|
|
chr5_-_121440704
|
0.695
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr7_+_101247601
|
0.694
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr7_-_134845363
|
0.693
|
NM_001008225
NM_001190847
NM_001190848
NM_001190849
NM_001190850
NM_013316
|
CNOT4
|
CCR4-NOT transcription complex, subunit 4
|
|
chr9_+_36026900
|
0.691
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr9_-_109291866
|
0.690
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr8_+_70567581
|
0.682
|
NM_001128205
NM_001128206
|
SULF1
|
sulfatase 1
|
|
chr3_-_187309442
|
0.681
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr20_+_57729659
|
0.679
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr3_-_162305319
|
0.679
|
NM_033169
NM_001038628
NM_003781
NM_033167
NM_033168
|
B3GALNT1
|
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group)
|
|
chr11_-_65871592
|
0.678
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr20_-_60074237
|
0.660
|
NM_003185
|
TAF4
|
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa
|
|
chr18_-_51406841
|
0.652
|
NM_001083962
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr2_+_178767317
|
0.645
|
NM_032523
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr1_-_95165089
|
0.644
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr20_-_48163071
|
0.643
|
NM_001032288
|
UBE2V1
|
ubiquitin-conjugating enzyme E2 variant 1
|
|
chr16_+_48616593
|
0.642
|
NM_153261
|
TMEM188
|
transmembrane protein 188
|
|
chr12_+_54612055
|
0.641
|
NM_201444
NM_201445
NM_201554
|
DGKA
|
diacylglycerol kinase, alpha 80kDa
|
|
chr21_-_14677310
|
0.636
|
NM_006948
|
HSPA13
|
heat shock protein 70kDa family, member 13
|
|
chr14_+_70443864
|
0.632
|
NM_014982
|
PCNX
|
pecanex homolog (Drosophila)
|
|
chr7_+_28305464
|
0.631
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_-_11838260
|
0.628
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr17_-_46553083
|
0.625
|
NM_001130528
NM_003971
|
SPAG9
|
sperm associated antigen 9
|
|
chr6_-_154873396
|
0.620
|
NM_173515
|
CNKSR3
|
CNKSR family member 3
|
|
chr8_-_82186832
|
0.616
|
NM_018440
|
PAG1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr3_-_15875932
|
0.616
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr8_-_49996353
|
0.613
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr10_-_61570665
|
0.611
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr1_+_23990228
|
0.608
|
NM_007260
|
LYPLA2
|
lysophospholipase II
|
|
chr6_-_24827265
|
0.600
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr13_+_92677012
|
0.598
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr5_-_14924555
|
0.597
|
NM_054027
|
ANKH
|
ankylosis, progressive homolog (mouse)
|
|
chr2_-_216009015
|
0.595
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr14_+_30098079
|
0.594
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr18_+_41560915
|
0.594
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr21_-_38792173
|
0.593
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr2_+_32142083
|
0.589
|
NM_014946
NM_199436
|
SPAST
|
spastin
|
|
chr2_-_142605739
|
0.588
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr10_-_126706442
|
0.585
|
NM_022802
|
CTBP2
|
C-terminal binding protein 2
|
|
chr16_+_48744310
|
0.585
|
NM_001040284
NM_001040285
|
PAPD5
|
PAP associated domain containing 5
|
|
chr2_-_19421852
|
0.582
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr8_-_93184629
|
0.582
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr3_-_69518119
|
0.580
|
NM_015123
|
FRMD4B
|
FERM domain containing 4B
|
|
chr12_+_5473558
|
0.579
|
NM_002527
|
NTF3
|
neurotrophin 3
|
|
chr6_+_47732181
|
0.579
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr17_+_5126275
|
0.572
|
NM_001083585
NM_004703
|
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1
|
|
chr20_-_4930133
|
0.566
|
NM_005116
|
SLC23A2
|
solute carrier family 23 (nucleobase transporters), member 2
|
|
chr1_+_178190493
|
0.558
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr13_-_32757835
|
0.558
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr14_-_38642165
|
0.556
|
NM_006364
|
SEC23A
|
Sec23 homolog A (S. cerevisiae)
|
|
chr6_+_148705421
|
0.550
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr2_-_156897286
|
0.549
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr2_+_11804190
|
0.545
|
NM_145693
|
LPIN1
|
lipin 1
|
|
chr15_+_45797977
|
0.545
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr2_-_202215593
|
0.539
|
NM_152388
|
ALS2CR4
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4
|
|
chr3_+_161040343
|
0.534
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr4_-_6434473
|
0.532
|
NM_181876
|
PPP2R2C
|
protein phosphatase 2, regulatory subunit B, gamma
|
|
chr6_-_57194977
|
0.532
|
NM_016277
|
RAB23
|
RAB23, member RAS oncogene family
|
|
chr14_+_58174489
|
0.518
|
NM_001079520
NM_016651
|
DACT1
|
dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis)
|
|
chr6_+_37333484
|
0.510
|
NM_017772
|
TBC1D22B
|
TBC1 domain family, member 22B
|
|
chr9_+_127550298
|
0.504
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr8_-_93181091
|
0.501
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_+_87921980
|
0.501
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chr14_-_69953433
|
0.500
|
NM_018373
|
SYNJ2BP
|
synaptojanin 2 binding protein
|