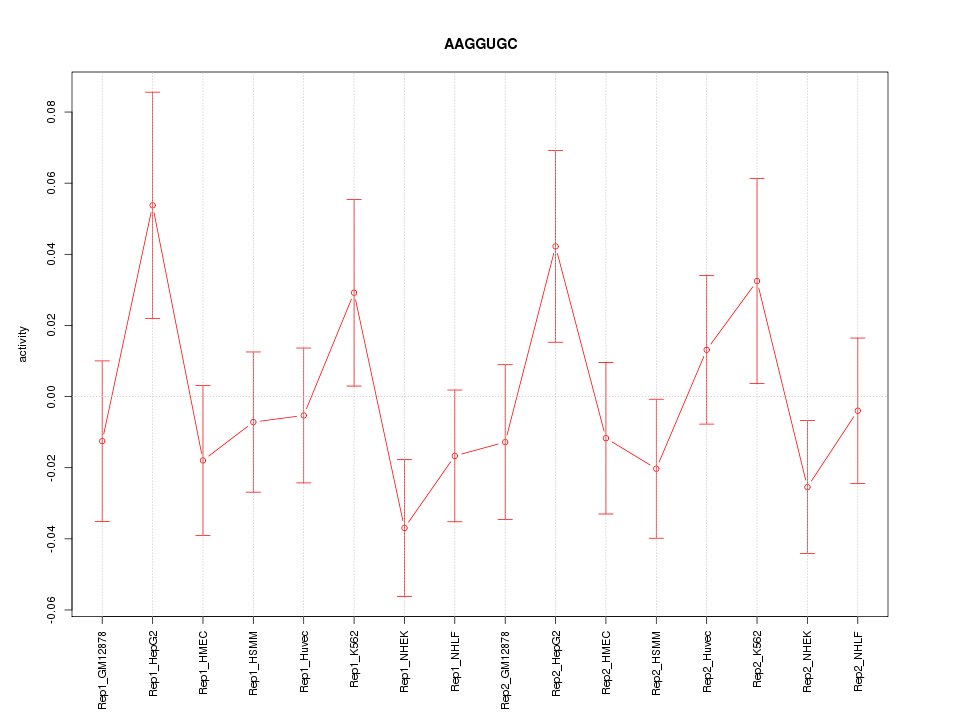
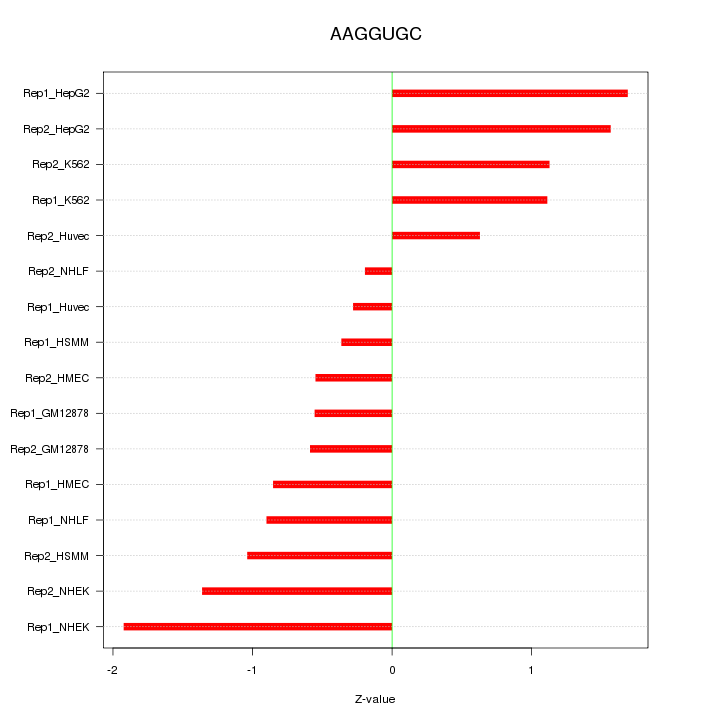
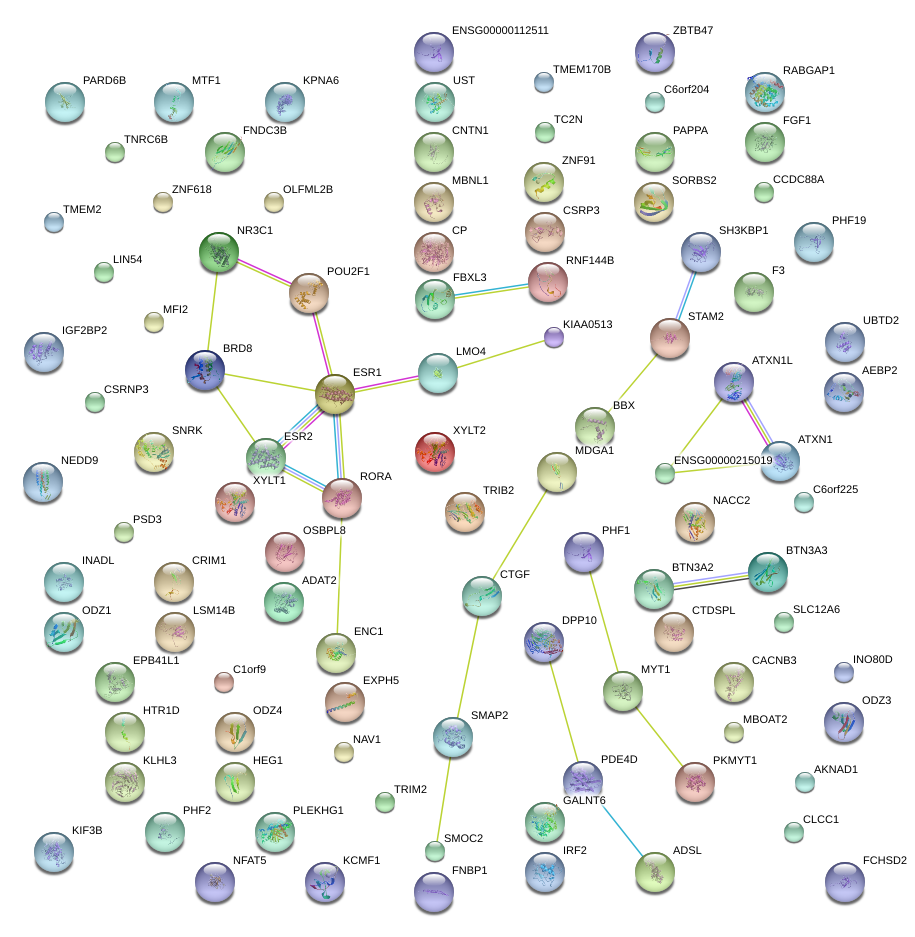
|
chr16_-_17472238
|
2.481
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr6_-_119137923
|
2.279
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr5_-_142795269
|
1.830
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr6_+_152053323
|
1.507
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr3_-_126257425
|
1.442
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr2_-_55500450
|
1.434
|
NM_001135597
NM_018084
|
CCDC88A
|
coiled-coil domain containing 88A
|
|
chr4_-_84152985
|
1.402
|
NM_001115007
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr3_+_37878647
|
1.347
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr17_+_45778381
|
1.288
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr1_-_109307539
|
1.245
|
NM_001048210
NM_015127
|
CLCC1
AKNAD1
|
chloride channel CLIC-like 1
AKNA domain containing 1
|
|
chr2_-_152740387
|
1.225
|
NM_005843
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr11_-_107969554
|
1.142
|
NM_015065
|
EXPH5
|
exophilin 5
|
|
chr2_-_206659140
|
1.103
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr20_+_34163671
|
1.040
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr15_-_32398220
|
1.034
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr5_-_73973004
|
0.970
|
NM_003633
|
ENC1
|
ectodermal-neural cortex 1 (with BTB-like domain)
|
|
chr3_+_42670179
|
0.966
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr20_+_30329096
|
0.943
|
NM_004798
|
KIF3B
|
kinesin family member 3B
|
|
chr16_+_70437394
|
0.937
|
NM_001137675
|
ATXN1L
|
ataxin 1-like
|
|
chr9_-_73573121
|
0.906
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr2_-_9061193
|
0.873
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr1_+_165456572
|
0.858
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr1_+_165564904
|
0.856
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr12_+_47498778
|
0.819
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr5_-_142046175
|
0.815
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr6_-_37773609
|
0.791
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr1_+_40612891
|
0.786
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chr3_+_43303005
|
0.773
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr6_+_11646472
|
0.769
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr22_+_38903825
|
0.758
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr3_+_153499883
|
0.749
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr1_-_160260210
|
0.728
|
NM_015441
|
OLFML2B
|
olfactomedin-like 2B
|
|
chr1_+_32346206
|
0.723
|
NM_012316
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7)
|
|
chr20_+_48781457
|
0.721
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr9_-_122679387
|
0.714
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr4_+_183482130
|
0.710
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr6_-_16869579
|
0.691
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr3_+_108724472
|
0.681
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr5_-_142057692
|
0.679
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr11_-_72530605
|
0.678
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr5_-_59819642
|
0.675
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_+_12774264
|
0.664
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr3_-_187025501
|
0.664
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr1_+_199975563
|
0.644
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr6_-_143813481
|
0.641
|
NM_182503
|
ADAT2
|
adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae)
|
|
chr5_-_142045770
|
0.636
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr2_+_166034402
|
0.629
|
NM_001172173
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr1_-_38097811
|
0.627
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr6_+_18495559
|
0.627
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr1_+_170768914
|
0.614
|
NM_016227
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr12_+_19483578
|
0.609
|
NM_001114176
NM_153207
|
AEBP2
|
AE binding protein 2
|
|
chr2_+_85051687
|
0.605
|
NM_020122
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chrX_-_19815586
|
0.583
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr6_+_150962691
|
0.559
|
NM_001029884
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1
|
|
chr1_-_23393808
|
0.556
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr9_+_124742932
|
0.546
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr6_+_112515366
|
0.533
|
NM_001033564
|
C6orf225
|
chromosome 6 open reading frame 225
|
|
chr9_+_117955890
|
0.521
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr4_-_186969041
|
0.520
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_87566738
|
0.506
|
NM_006769
|
LMO4
|
LIM domain only 4
|
|
chr9_+_95378492
|
0.501
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr1_+_40635058
|
0.497
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr9_-_138126917
|
0.495
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr6_-_132314004
|
0.470
|
NM_001901
|
CTGF
|
connective tissue growth factor
|
|
chr8_-_18915443
|
0.453
|
NM_015310
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr3_+_173240983
|
0.427
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr20_+_60130866
|
0.426
|
NM_144703
|
LSM14B
|
LSM14B, SCD6 homolog B (S. cerevisiae)
|
|
chr5_-_141973875
|
0.425
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr1_-_94779727
|
0.417
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr15_-_59308770
|
0.408
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr9_+_115678365
|
0.405
|
NM_133374
|
ZNF618
|
zinc finger protein 618
|
|
chr5_-_137099602
|
0.404
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr5_-_171643399
|
0.393
|
NM_152277
|
UBTD2
|
ubiquitin domain containing 2
|
|
chr19_-_23370066
|
0.392
|
NM_003430
|
ZNF91
|
zinc finger protein 91
|
|
chr13_-_76499297
|
0.389
|
NM_012158
|
FBXL3
|
F-box and leucine-rich repeat protein 3
|
|
chr15_-_58671929
|
0.389
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr16_+_83618804
|
0.389
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr6_-_119079712
|
0.384
|
NM_001042475
NM_206921
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr2_+_166137091
|
0.380
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr3_-_150422474
|
0.378
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr4_-_185632610
|
0.374
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr15_-_58706934
|
0.365
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr1_+_170768635
|
0.363
|
NM_014283
|
C1orf9
|
chromosome 1 open reading frame 9
|
|
chr6_-_11340887
|
0.348
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr4_+_154293719
|
0.345
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr4_-_186969202
|
0.345
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_61980679
|
0.342
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr16_+_68156497
|
0.341
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr2_+_36436873
|
0.339
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr4_+_154345039
|
0.334
|
NM_015271
|
TRIM2
|
tripartite motif containing 2
|
|
chr2_+_114935623
|
0.332
|
NM_001178036
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr6_+_149109841
|
0.329
|
NM_005715
|
UST
|
uronyl-2-sulfotransferase
|
|
chr9_-_131845277
|
0.326
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr11_-_78829342
|
0.322
|
NM_001098816
|
ODZ4
|
odz, odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr6_+_26473376
|
0.318
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr6_+_138230045
|
0.315
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr6_-_111243100
|
0.315
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr1_-_120413798
|
0.313
|
NM_024408
|
NOTCH2
|
notch 2
|
|
chr5_+_31834787
|
0.312
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr11_+_109469296
|
0.310
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr2_+_63131466
|
0.305
|
NM_014562
|
OTX1
|
orthodenticle homeobox 1
|
|
chr2_-_179051471
|
0.302
|
NM_001135212
NM_181342
|
FKBP7
|
FK506 binding protein 7
|
|
chr12_-_54869568
|
0.302
|
NM_001130420
NM_003075
NM_139067
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr7_+_117611321
|
0.294
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr20_+_226200
|
0.292
|
NM_033089
|
ZCCHC3
|
zinc finger, CCHC domain containing 3
|
|
chr17_+_24742068
|
0.288
|
NM_020791
|
TAOK1
|
TAO kinase 1
|
|
chr12_-_120218926
|
0.283
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr11_+_94462585
|
0.280
|
NM_015036
|
ENDOD1
|
endonuclease domain containing 1
|
|
chr6_-_143307976
|
0.279
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr15_+_62991106
|
0.276
|
NM_182703
|
ANKDD1A
|
ankyrin repeat and death domain containing 1A
|
|
chr18_-_43710917
|
0.275
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr7_-_120823591
|
0.273
|
NM_001040020
NM_014888
|
FAM3C
|
family with sequence similarity 3, member C
|
|
chr4_+_24923490
|
0.270
|
NM_024936
|
ZCCHC4
|
zinc finger, CCHC domain containing 4
|
|
chr20_+_51022352
|
0.262
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr6_-_42527760
|
0.261
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr20_+_43028533
|
0.260
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr7_+_66023564
|
0.254
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr18_-_31124141
|
0.253
|
NM_001112734
NM_001166012
|
ZSCAN30
|
zinc finger and SCAN domain containing 30
|
|
chr14_+_30098079
|
0.248
|
NM_017769
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase
|
|
chr20_-_43410437
|
0.246
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr4_-_48477038
|
0.245
|
NM_015030
|
FRYL
|
FRY-like
|
|
chr1_-_62926533
|
0.243
|
NM_033407
|
DOCK7
|
dedicator of cytokinesis 7
|
|
chr4_+_108965167
|
0.236
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr12_+_54646814
|
0.236
|
NM_001798
NM_052827
|
CDK2
|
cyclin-dependent kinase 2
|
|
chr16_-_20660618
|
0.234
|
NM_017736
|
THUMPD1
|
THUMP domain containing 1
|
|
chr5_-_158567411
|
0.234
|
NM_144726
|
RNF145
|
ring finger protein 145
|
|
chr12_+_4253143
|
0.233
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr6_+_47732181
|
0.228
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr14_+_31616196
|
0.226
|
NM_001030055
NM_001173
|
ARHGAP5
|
Rho GTPase activating protein 5
|
|
chr17_+_76988134
|
0.226
|
NM_001080519
|
BAHCC1
|
BAH domain and coiled-coil containing 1
|
|
chr1_+_144149794
|
0.224
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr12_+_108020305
|
0.223
|
NM_003362
|
UNG
|
uracil-DNA glycosylase
|
|
chr6_+_28425666
|
0.220
|
NM_024493
|
ZKSCAN3
|
zinc finger with KRAB and SCAN domains 3
|
|
chr16_+_84202515
|
0.217
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr2_+_201691513
|
0.217
|
NM_001127183
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr5_-_141981083
|
0.215
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr19_+_21056792
|
0.215
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr17_+_65677255
|
0.215
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr20_-_5041482
|
0.214
|
NM_001009923
NM_001009924
NM_001009925
NM_014145
|
C20orf30
|
chromosome 20 open reading frame 30
|
|
chr2_-_101370355
|
0.214
|
NM_153836
|
CREG2
|
cellular repressor of E1A-stimulated genes 2
|
|
chr2_+_32244406
|
0.213
|
NM_001193513
NM_001193514
NM_001193515
NM_017964
|
SLC30A6
|
solute carrier family 30 (zinc transporter), member 6
|
|
chr17_+_38719371
|
0.212
|
NM_145041
|
TMEM106A
FLJ77644
|
transmembrane protein 106A
hypothetical LOC728772
|
|
chr6_+_122762384
|
0.211
|
NM_001135564
NM_004506
|
HSF2
|
heat shock transcription factor 2
|
|
chr10_-_71600191
|
0.209
|
NM_001142648
NM_020150
|
SAR1A
|
SAR1 homolog A (S. cerevisiae)
|
|
chr15_-_32416336
|
0.209
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr2_+_75727415
|
0.207
|
NM_014763
|
MRPL19
|
mitochondrial ribosomal protein L19
|
|
chr17_+_4972410
|
0.205
|
NM_004505
|
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene)
|
|
chr10_-_98263434
|
0.202
|
NM_012465
|
TLL2
|
tolloid-like 2
|
|
chr2_-_179837490
|
0.202
|
NM_178123
|
SESTD1
|
SEC14 and spectrin domains 1
|
|
chr1_+_161308318
|
0.202
|
NM_001113380
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr22_+_29420792
|
0.196
|
NM_030758
|
OSBP2
|
oxysterol binding protein 2
|
|
chr8_-_18710575
|
0.196
|
NM_206909
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr4_+_44375165
|
0.195
|
NM_021927
|
GUF1
|
GUF1 GTPase homolog (S. cerevisiae)
|
|
chr2_+_173648751
|
0.194
|
NM_016653
NM_133646
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr21_-_35182842
|
0.193
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr6_-_49538810
|
0.190
|
NM_000255
|
MUT
|
methylmalonyl CoA mutase
|
|
chr3_+_106568334
|
0.189
|
NM_001627
|
ALCAM
|
activated leukocyte cell adhesion molecule
|
|
chr12_-_54938385
|
0.189
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr20_+_42537927
|
0.189
|
NM_001039199
NM_024331
|
TTPAL
|
tocopherol (alpha) transfer protein-like
|
|
chr2_-_130603264
|
0.188
|
NM_001099771
|
POTEF
|
POTE ankyrin domain family, member F
|
|
chr4_-_42353640
|
0.188
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr8_-_81246215
|
0.184
|
NM_001025253
NM_005079
|
TPD52
|
tumor protein D52
|
|
chr16_-_55777452
|
0.182
|
NM_024946
|
FAM192A
|
family with sequence similarity 192, member A
|
|
chr7_+_44754978
|
0.181
|
NM_031449
|
ZMIZ2
|
zinc finger, MIZ-type containing 2
|
|
chr2_+_171493870
|
0.180
|
NM_015530
|
GORASP2
|
golgi reassembly stacking protein 2, 55kDa
|
|
chr4_+_167013859
|
0.180
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr7_-_111633620
|
0.178
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr1_-_43606276
|
0.178
|
NM_022821
|
ELOVL1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
|
|
chr11_-_16454493
|
0.175
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr22_+_38770610
|
0.173
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr6_+_31016755
|
0.170
|
NM_080870
|
DPCR1
|
diffuse panbronchiolitis critical region 1
|
|
chr12_+_102982365
|
0.168
|
NM_013320
|
HCFC2
|
host cell factor C2
|
|
chr18_+_53865455
|
0.168
|
NM_001144964
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr16_+_30867878
|
0.167
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr5_+_56507029
|
0.167
|
NM_001127235
|
GPBP1
|
GC-rich promoter binding protein 1
|
|
chr6_+_31190505
|
0.166
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr20_+_56318155
|
0.165
|
NM_020673
|
RAB22A
|
RAB22A, member RAS oncogene family
|
|
chr9_-_98220482
|
0.162
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr2_+_112749617
|
0.160
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr4_-_186970366
|
0.160
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr14_-_94669547
|
0.159
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr6_+_26548678
|
0.153
|
NM_006994
NM_197974
|
BTN3A3
|
butyrophilin, subfamily 3, member A3
|
|
chr6_+_26705110
|
0.153
|
NM_013375
|
ABT1
|
activator of basal transcription 1
|
|
chr18_-_3001944
|
0.151
|
NM_014646
|
LPIN2
|
lipin 2
|
|
chr1_+_117711575
|
0.149
|
NM_006699
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2
|
|
chr3_-_170348215
|
0.148
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr7_-_102576570
|
0.148
|
NM_001122838
NM_198990
|
NAPEPLD
|
N-acyl phosphatidylethanolamine phospholipase D
|
|
chr6_-_24491498
|
0.147
|
NM_001195610
|
DCDC2
|
doublecortin domain containing 2
|
|
chr4_+_158216631
|
0.144
|
NM_000824
NM_001166061
NM_001166060
|
GLRB
|
glycine receptor, beta
|
|
chr10_+_126620681
|
0.144
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chrX_-_19727828
|
0.143
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_32777358
|
0.143
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr17_-_7323667
|
0.143
|
NM_001128833
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr17_-_59694384
|
0.140
|
NM_018469
|
TEX2
|
testis expressed 2
|
|
chr1_-_152162074
|
0.140
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr10_+_116687904
|
0.138
|
NM_139169
|
TRUB1
|
TruB pseudouridine (psi) synthase homolog 1 (E. coli)
|
|
chr22_-_18687511
|
0.138
|
NM_033257
|
DGCR6L
|
DiGeorge syndrome critical region gene 6-like
|
|
chr11_+_32870935
|
0.136
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr1_-_204048758
|
0.136
|
NM_173854
|
SLC41A1
|
solute carrier family 41, member 1
|
|
chr6_+_30647148
|
0.136
|
NM_001025091
NM_001090
|
ABCF1
|
ATP-binding cassette, sub-family F (GCN20), member 1
|